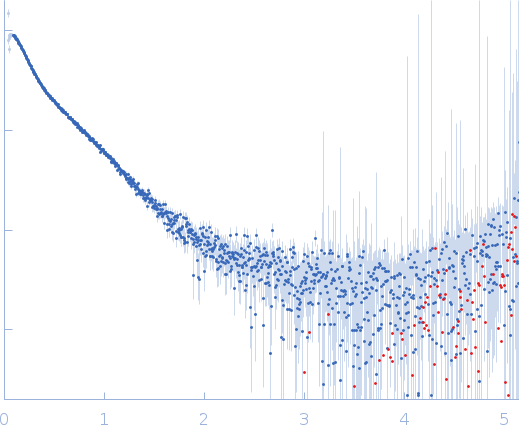
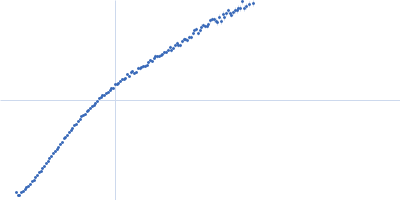
The disordered C-terminal tail of fungal LPMOs from phytopathogens mediates protein dimerization and impacts plant penetration.
Tamburrini KC,
Kodama S,
Grisel S,
Haon M,
Nishiuchi T,
Bissaro B,
Kubo Y,
Longhi S,
Berrin JG
Proc Natl Acad Sci U S A
121(13):e2319998121
(2024 Mar 26)
|
|
|
|
|
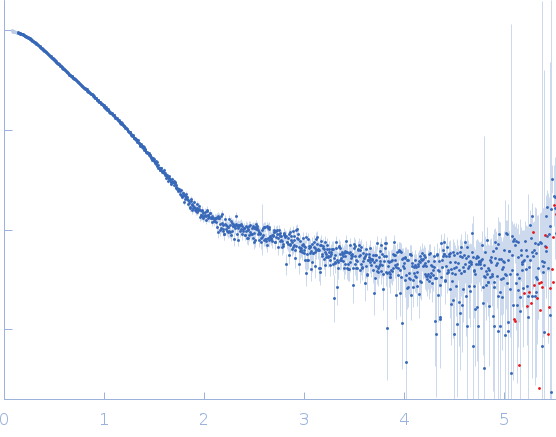
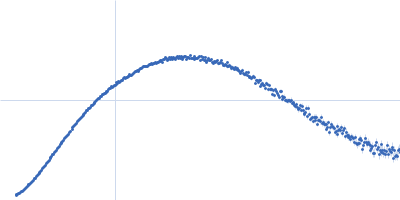
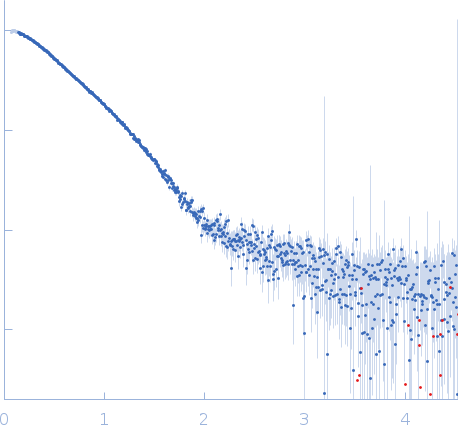
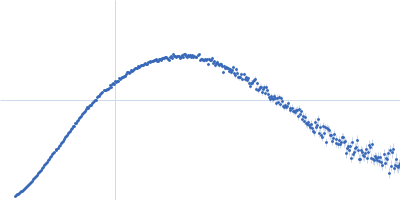
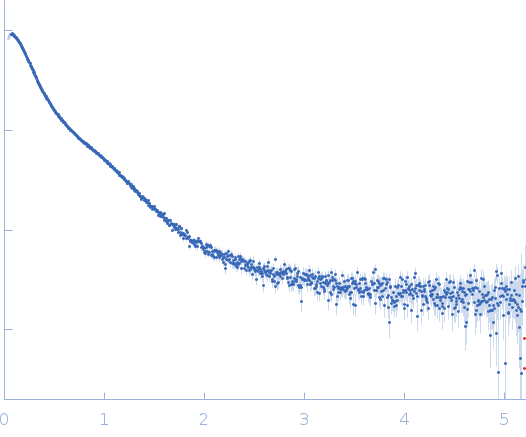
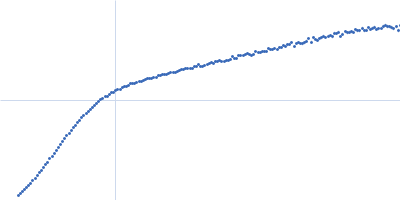
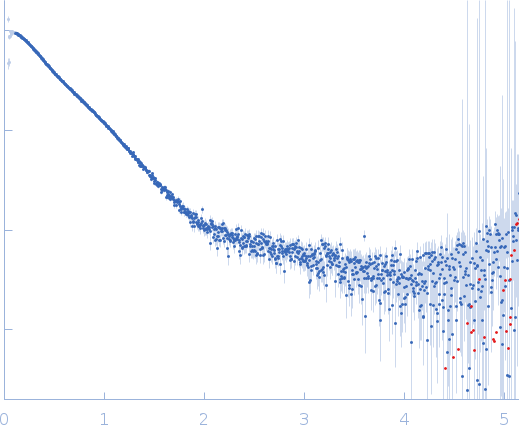
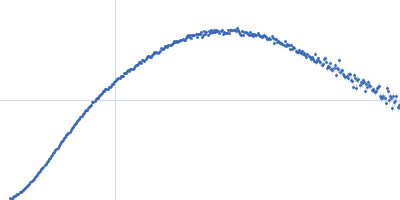
| Sample: |
Endoglucanase-7 monomer, 30 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
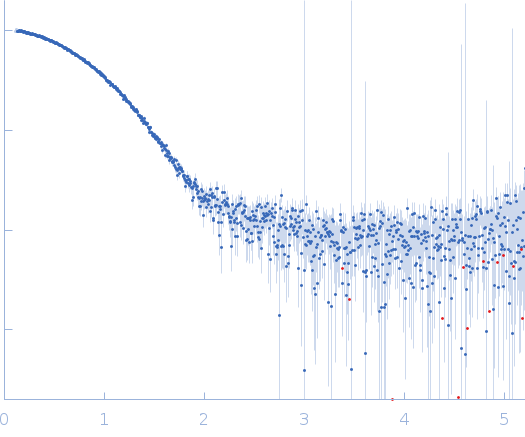
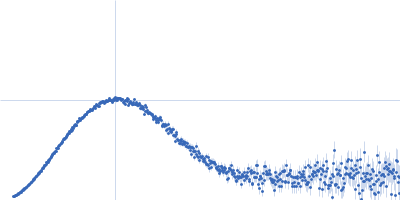
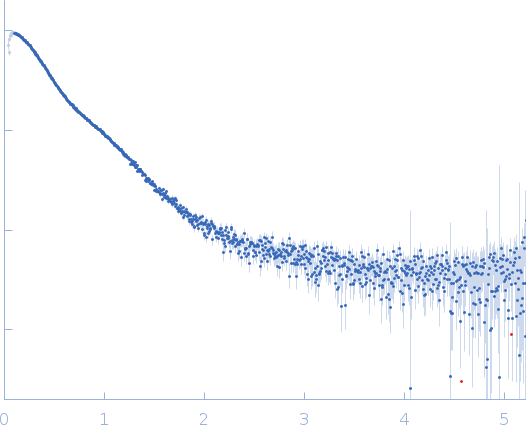
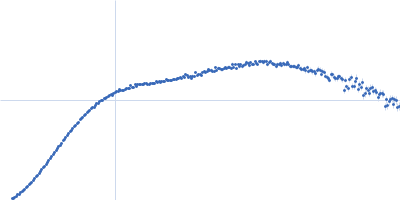
| Sample: |
Endoglucanase-7 dimer, 60 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 1
|
|
| RgGuinier |
4.8 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-7 monomer, 27 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Feb 1
|
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-7 (C286A) monomer, 30 kDa Colletotrichum orbiculare (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Feb 1
|
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glycosyl hydrolase family 61 monomer, 30 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 9
|
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glycosyl hydrolase family 61 dimer, 61 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 9
|
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-4 monomer, 33 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Feb 17
|
|
| RgGuinier |
3.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoglucanase-4 dimer, 66 kDa Colletotrichum higginsianum (strain … protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Feb 17
|
|
| RgGuinier |
6.4 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
199 |
nm3 |
|
|