Optimization of Structure‐Guided Development of Chemical Probes for the Pseudoknot RNA of the Frameshift Element in SARS‐CoV‐2
Ceylan B,
Adam J,
Toews S,
Kaiser F,
Dörr J,
Scheppa D,
Tants J,
Smart A,
Schoth J,
Philipp S,
Stirnal E,
Ferner J,
Richter C,
Sreeramulu S,
Caliskan N,
Schlundt A,
Weigand J,
Göbel M,
Wacker A,
Schwalbe H
Angewandte Chemie International Edition
(2025 Feb 05)
|
|
|
|
|
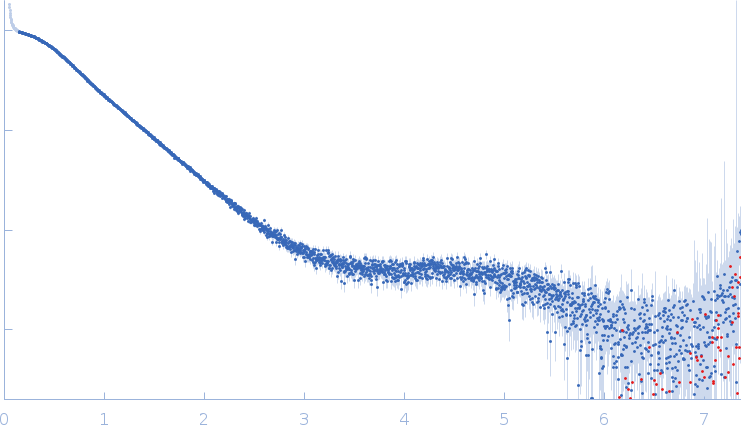
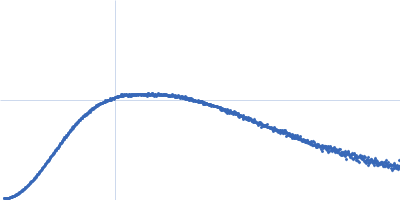
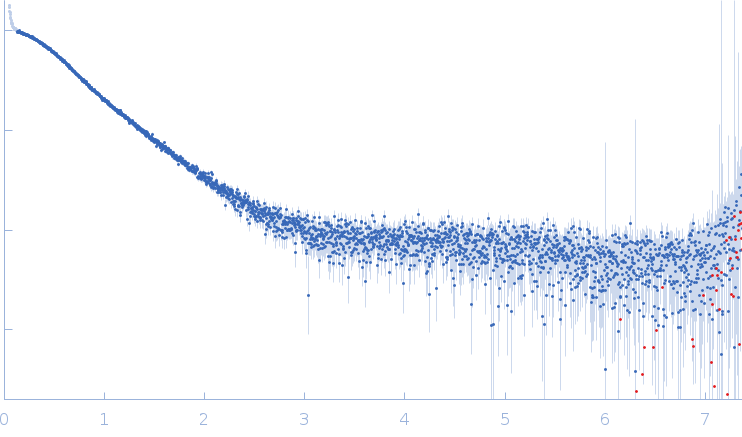
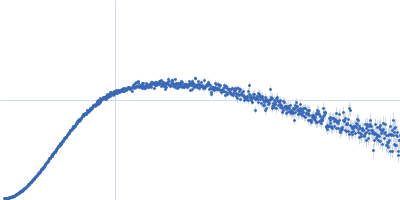
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
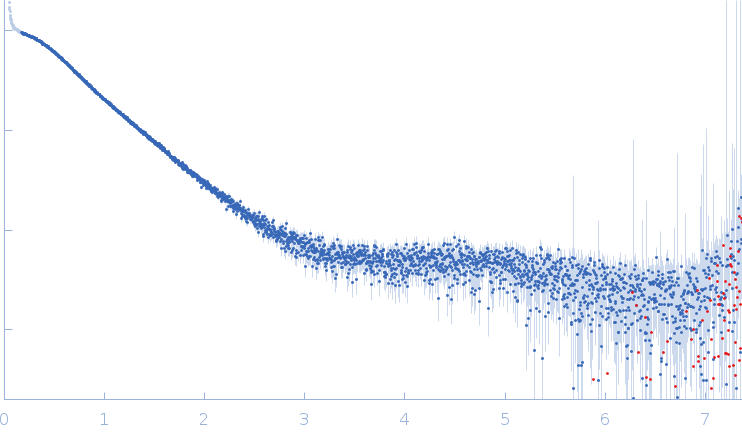
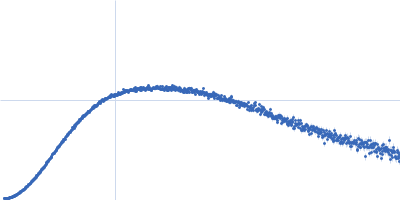
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|