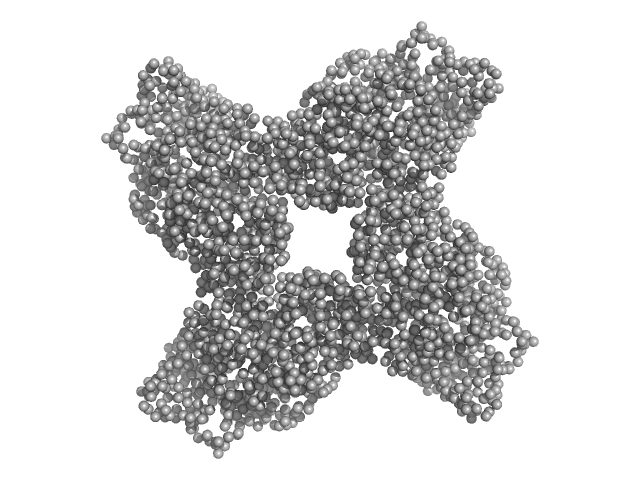
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Chandravanshi K,
Singh R,
Kumar A,
Bhange GN,
Kumar A,
Makde RD
Int J Biol Macromol
:136734
(2024 Oct 19)
|
|
|
|
|
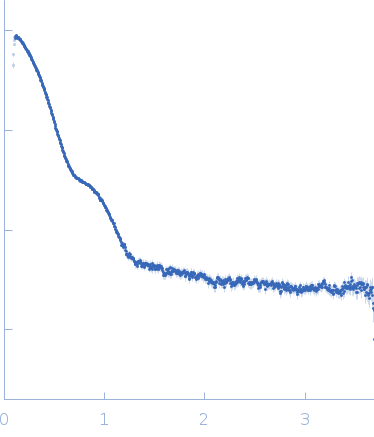
| Sample: |
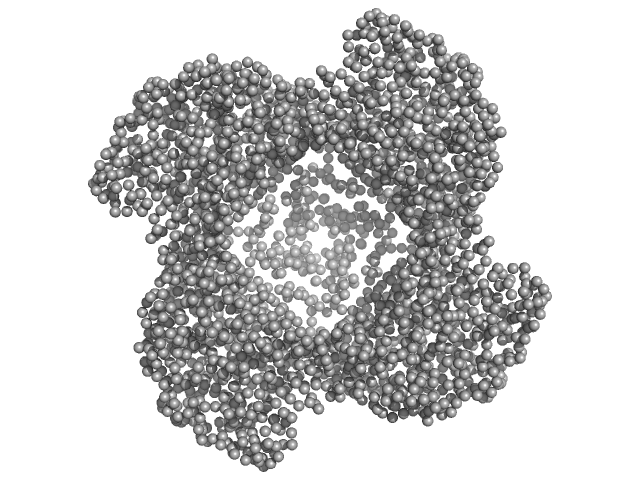
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
439 |
nm3 |
|
|
|
|
|
|
|
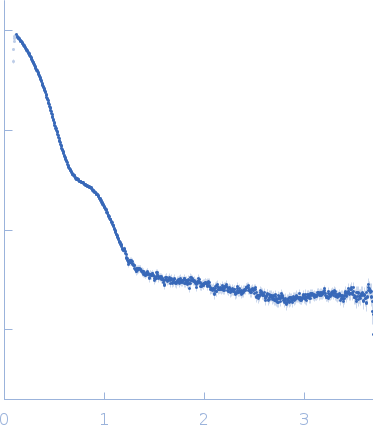
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
435 |
nm3 |
|
|
|
|
|
|

|
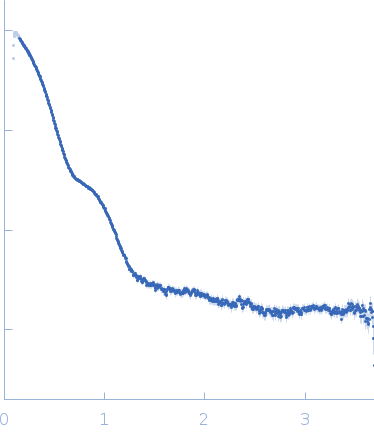
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
446 |
nm3 |
|
|