Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Gondelaud F,
Leval J,
Arora L,
Walimbe A,
Bignon C,
Ptchelkine D,
Brocca S,
Mukhopadyay S,
Longhi S
Protein Sci
34(3):e70068
(2025 Mar)
|
|
|
|
|
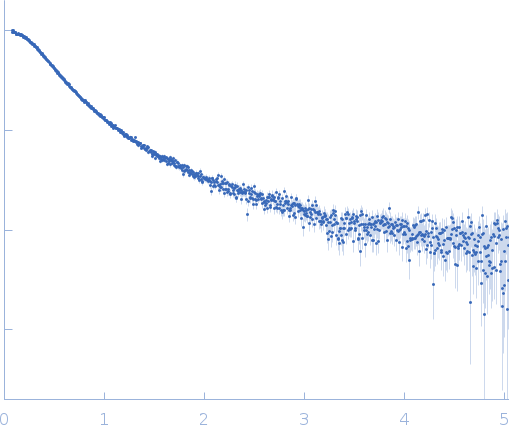
| Sample: |
Protein W monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
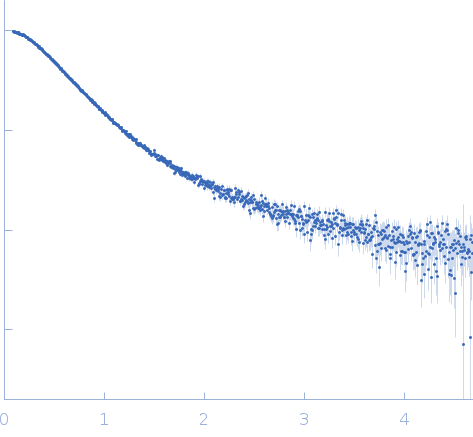
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
|
| RgGuinier |
3.4 |
nm |
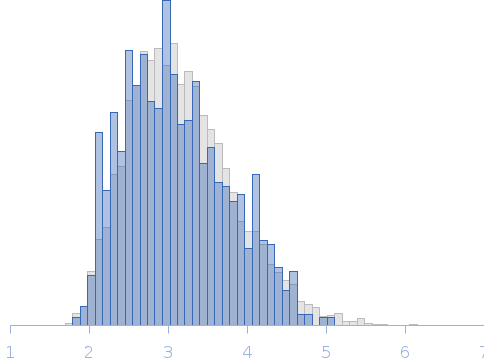
| Dmax |
13.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
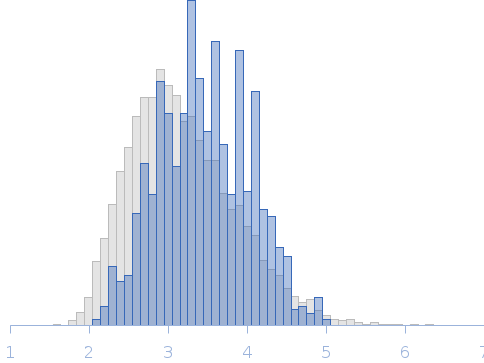
| Sample: |
Protein W (artificial PNT3 variant - low-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
|
| RgGuinier |
3.8 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
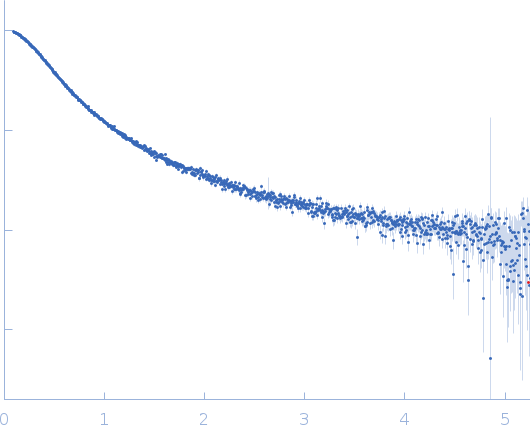
| Sample: |
Protein W (artificial PNT3 variant - high-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 5
|
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
54 |
nm3 |
|
|