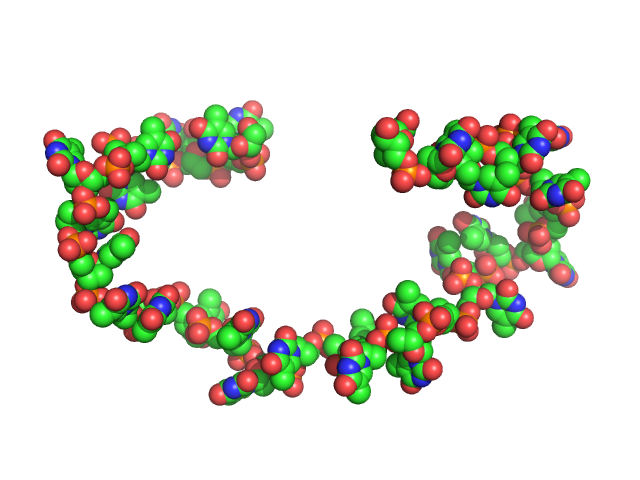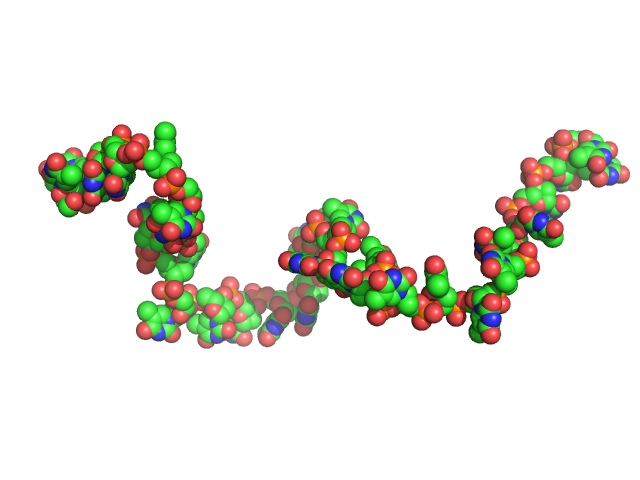The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Plumridge A,
Meisburger SP,
Andresen K,
Pollack L
Nucleic Acids Res
45(7):3932-3943
(2017 Apr 20)
|
|
|
|
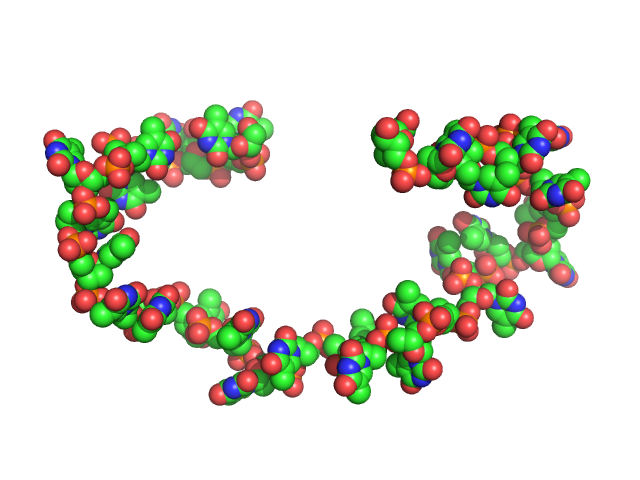
|
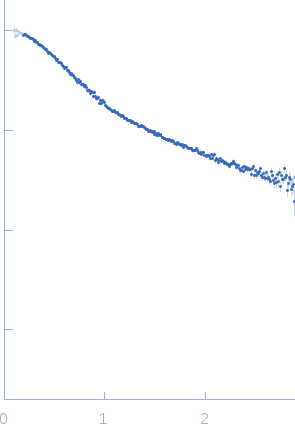
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 100mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
|
|
|
|
|
|
|
|
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 200mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
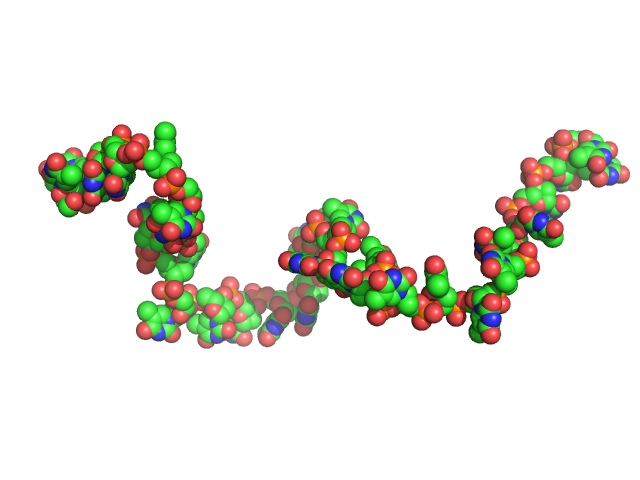
|
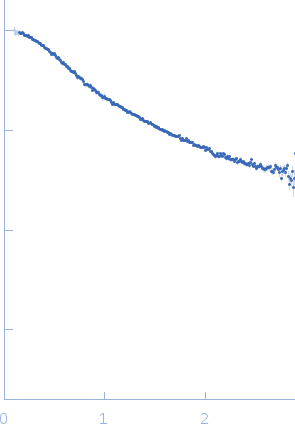
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 200mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
|
|
|
|
|
|
|
|
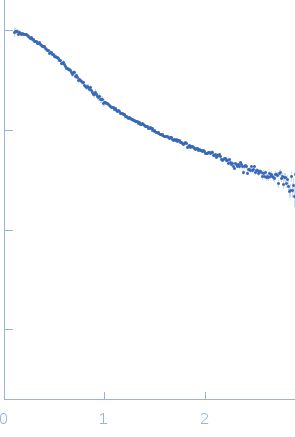
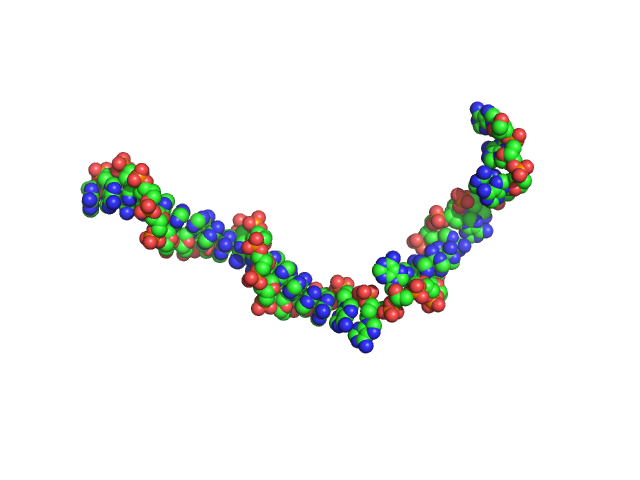
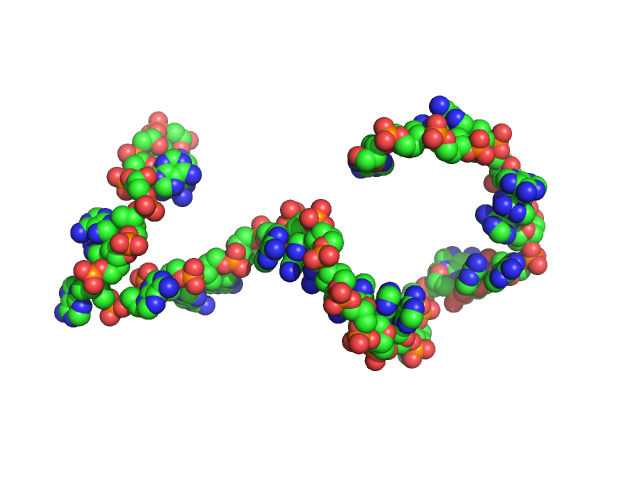
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 100mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|