A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Mardle CE,
Shakespeare TJ,
Butt LE,
Goddard LR,
Gowers DM,
Atkins HS,
Vincent HA,
Callaghan AJ
Sci Rep
9(1):7952
(2019 May 28)
|
|
|
|

|
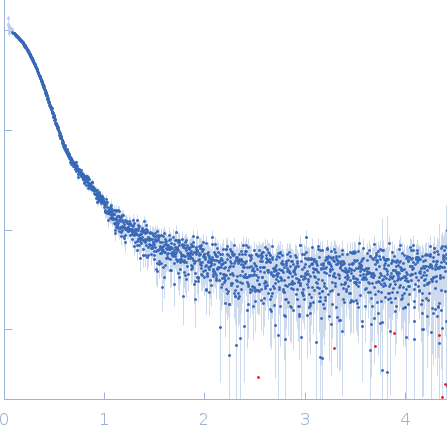
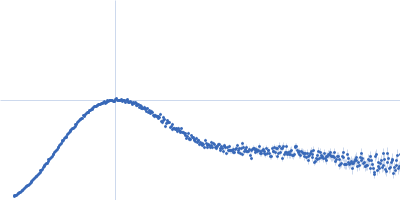
| Sample: |
Endoribonuclease E tetramer, 247 kDa Escherichia coli protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
468 |
nm3 |
|
|
|
|
|
|
|
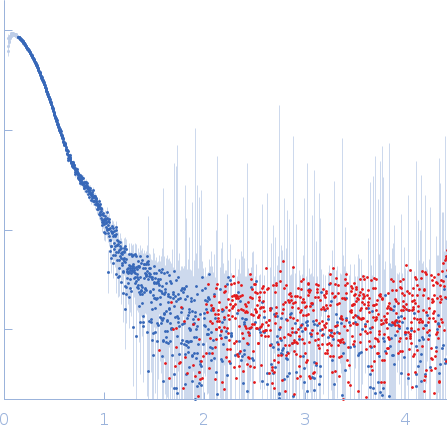
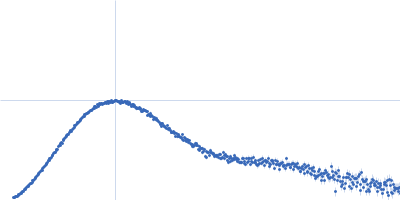
| Sample: |
Endoribonuclease E tetramer, 248 kDa Yersinia pestis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
470 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 256 kDa Francisella tularensis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
491 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 250 kDa Burkholderia pseudomallei protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 254 kDa Acinetobacter baumannii protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
508 |
nm3 |
|
|