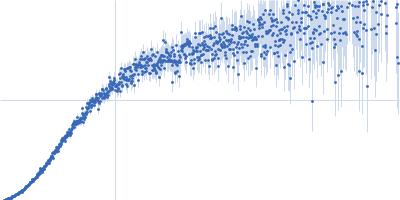
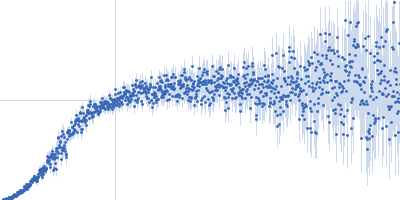
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Fuertes G,
Banterle N,
Ruff KM,
Chowdhury A,
Mercadante D,
Koehler C,
Kachala M,
Estrada Girona G,
Milles S,
Mishra A,
Onck PR,
Gräter F,
Esteban-Martín S,
Pappu RV,
Svergun DI,
Lemke EA
Proc Natl Acad Sci U S A
114(31):E6342-E6351
(2017 Aug 1)
|
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 8
|
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 8
|
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 8
|
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoporin NUP49/NSP49 monomer, 4 kDa Saccharomyces cerevisiae protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Inner nuclear membrane protein HEH2 monomer, 5 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 25
|
|
| RgGuinier |
2.4 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Inner nuclear membrane protein HEH2 monomer, 5 kDa Saccharomyces cerevisiae protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 25
|
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Inner nuclear membrane protein HEH2 monomer, 5 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 25
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Inner nuclear membrane protein HEH2 monomer, 5 kDa Saccharomyces cerevisiae protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 25
|
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Importin subunit alpha-1 monomer, 11 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 8
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Importin subunit alpha-1 monomer, 11 kDa Homo sapiens protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 9
|
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Importin subunit alpha-1 monomer, 11 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
3.1 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Importin subunit alpha-1 monomer, 11 kDa Homo sapiens protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 8 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 8 kDa Homo sapiens protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 8 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 8 kDa Homo sapiens protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 12 kDa Homo sapiens protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup153 monomer, 12 kDa Homo sapiens protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex monomer, 4 kDa Escherichia coli protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex monomer, 4 kDa Escherichia coli protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 8
|
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cold shock-like protein monomer, 7 kDa Thermotoga maritima protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 16
|
|
| RgGuinier |
2.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cold shock-like protein monomer, 7 kDa Thermotoga maritima protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Thioredoxin 1 monomer, 12 kDa Escherichia coli protein
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Thioredoxin 1 monomer, 12 kDa Escherichia coli protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup98-Nup96 monomer, 15 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 24
|
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoporin NSP1 monomer, 18 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 24
|
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
45 |
nm3 |
|
|