Inhibition of the Staphylococcus aureus c-di-AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM.
Tosi T,
Hoshiga F,
Millership C,
Singh R,
Eldrid C,
Patin D,
Mengin-Lecreulx D,
Thalassinos K,
Freemont P,
Gründling A
PLoS Pathog
15(1):e1007537
(2019 Jan 22)
|
|
|
|

|
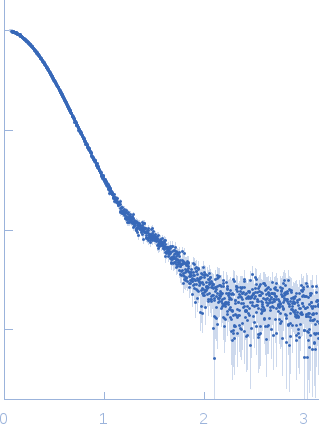
| Sample: |
Diadenylate cyclase dimer, 39 kDa Staphylococcus aureus protein
Phosphoglucosamine mutase dimer, 99 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|

|
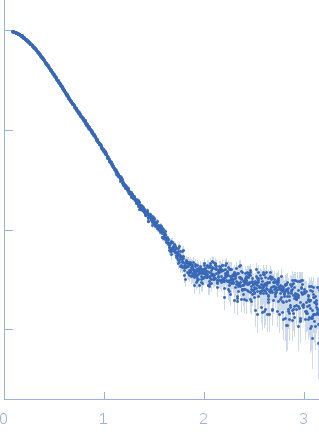
| Sample: |
Phosphoglucosamine mutase dimer, 99 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diadenylate cyclase dimer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 May 7
|
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
57 |
nm3 |
|
|