Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility in Escherichia coli.
Hou YJ,
Yang WS,
Hong Y,
Zhang Y,
Wang DC,
Li DF
J Biol Chem
295(3):808-821
(2020 Jan 17)
|
|
|
|

|
| Sample: |
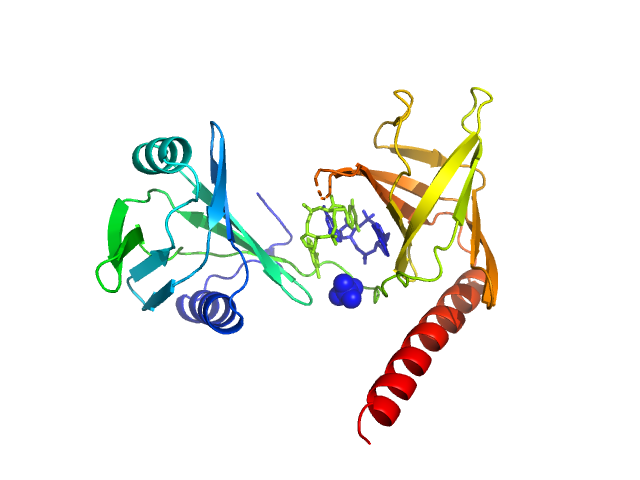
Flagellar brake protein YcgR monomer, 29 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150mM NaCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jan 4
|
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Flagellar brake protein YcgR in complex with c-di-GMP monomer, 29 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150mM NaCl, 10% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jan 4
|
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
44 |
nm3 |
|
|




