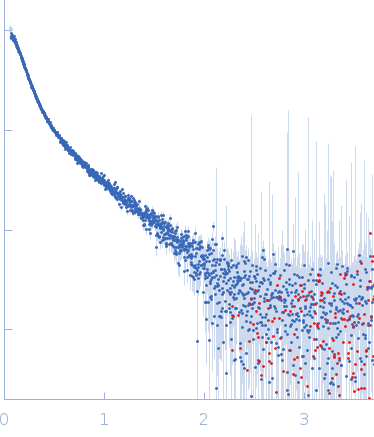
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Kim D,
Thiel B,
Mrozowich T,
Hennelly S,
Hofacker I,
Patel T,
Sanbonmatsu K
Nature Communications
11(1)
(2020 Jan 9)
|
|
|
|

|
| Sample: |
Braveheart Fragment 1 monomer, 116 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
|
| RgGuinier |
8.4 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
426 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart Fragment 2 monomer, 112 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
|
| RgGuinier |
8.1 |
nm |
| Dmax |
27.2 |
nm |
| VolumePorod |
443 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart Fragment 3 monomer, 111 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
|
| RgGuinier |
6.5 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
492 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
|
| RgGuinier |
9.9 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
2150 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
|
| RgGuinier |
11.8 |
nm |
| Dmax |
28.7 |
nm |
| VolumePorod |
2380 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 12 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jul 20
|
|
| RgGuinier |
9.7 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
1370 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart 5' Module monomer, 32 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart 5' Module monomer, 32 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 12 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
|
| RgGuinier |
4.8 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart 3' module monomer, 72 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
|
| RgGuinier |
6.5 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart 3' module monomer, 72 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 12 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 16
|
|
| RgGuinier |
7.3 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
|
| RgGuinier |
9.8 |
nm |
| Dmax |
30.2 |
nm |
| VolumePorod |
1660 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
Braveheart Fragment 1 monomer, 116 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
|
| RgGuinier |
8.2 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
455 |
nm3 |
|
|