|
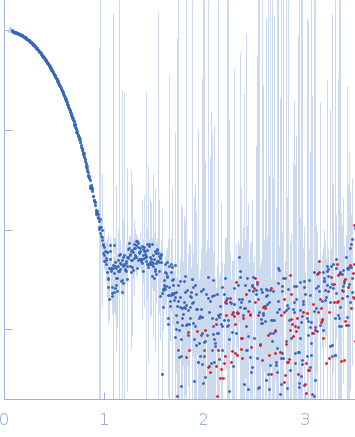
Synchrotron SAXS data from solutions of Glucose Isomerase in PBS pH 7.4 were collected using size exclusion chromatography coupled SAXS (SEC-SAXS) on the BioSAXS Beamline at the Australian Synchrotron ANSTO. A Pilatus3X 2M detector was used with a sample-to-detector distance of 4.1 m and a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). 50 µl of glucose isomerase at 3.8 mg/ml was passed through a Superdex 200 5/150 Increase chromatography column (Cytiva) at a flow rate of 0.4 mg/ml and eluted directly into a capillary with a flowing sheath fluid of PBS buffer. Scattering images of the flowing solution in the capillary were acquired with 1 s exposure times per frame for the duration of the size exclusion column. The data were normalized to the intensity of the transmitted beam and radially averaged. Initial acquisitions of the buffer only were averaged and subtracted from the acquisitions taken during protein elution to generate the background-subtracted sample scattering profiles.
|
|
Xylose isomerase
|
| Mol. type |
|
Protein |
| Organism |
|
Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) |
| Olig. state |
|
Tetramer |
| Mon. MW |
|
48.9 kDa |
| |
| UniProt |
|
Q8A9M2
(1-438)
|
| Sequence |
|
FASTA |
| |
|
 s, nm-1
s, nm-1
