|
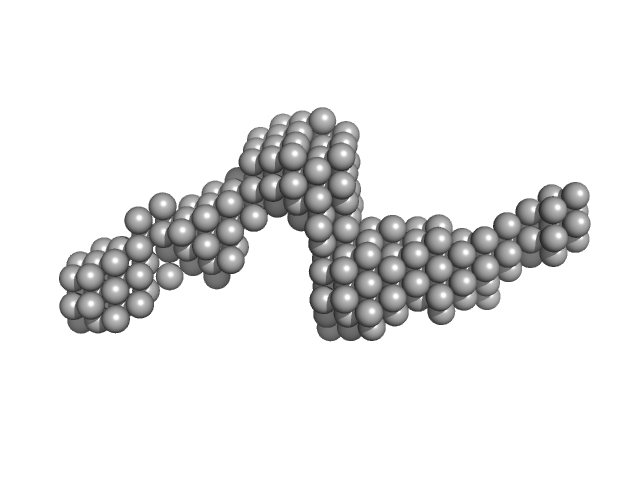
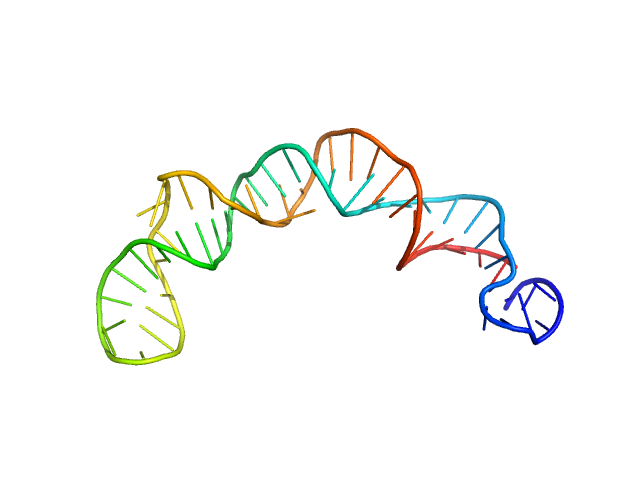
SAXS data for the Gli-55 aptamer were obtained at the P12 BioSAXS beamline (EMBL) at the Petra III storage ring of the synchrotron center DESY, Hamburg. The eluent of the employed chromatography column was passed through a 1.7 mm quartz capillary held under vacuum. The SAXS data were recorded on a Pilatus 6M area detector (Dectris) at a sample to detector distance of 3 meters and the wavelength λ = 0.124 nm (X-ray energy 10 keV) at room temperature 20.4 °C. Series of individual 1 s exposure X-ray data frames were measured from the continuously-flowing column eluate across one column volume. The two-dimensional SAXS intensities were reduced to I(s) vs s using the integrated analysis pipeline SASFLOW. The s-axis was calibrated with silver behenate and the resulting profiles were normalized for exposure time and sample transmission. To distinguish the oligomeric constituents in solution the joint technique of size-exclusion chromatography with SAXS (SEC-SAXS) was applied. To decompose the partially overlapping components in SEC-SAXS data the evolving factor analysis (EFA) was applied using the program EFAMIX.
|
|
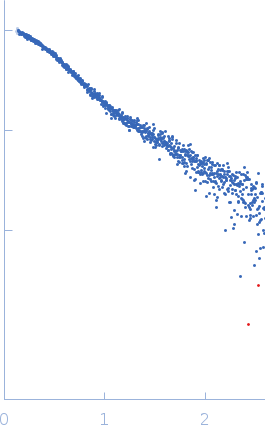 s, nm-1
s, nm-1