|
|
|
|
|
| Sample: |
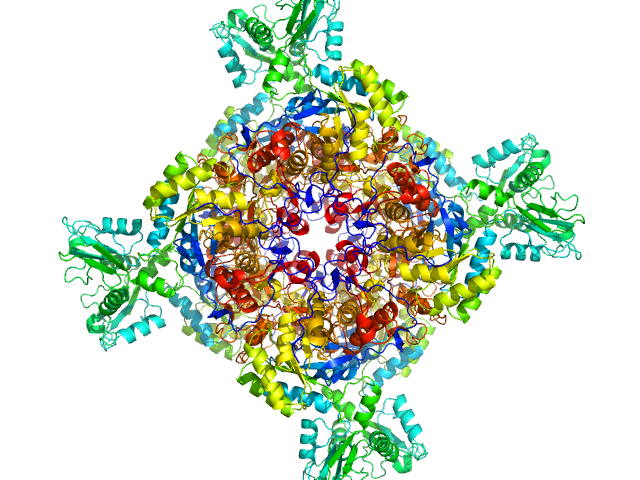
Inosine-5'-monophosphate dehydrogenase octamer, 426 kDa Mycolicibacterium smegmatis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 2 mM MgCl2, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2024 Feb 21
|
...dehydrogenase
Nature Communications 15(1) (2024)
Bulvas O, Knejzlík Z, Sýs J, Filimoněnko A, Čížková M, Clarová K, Rejman D, Kouba T, Pichová I
|
| RgGuinier |
5.3 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
952 |
nm3 |
|