|
SAXS data from solutions of Mycobacterium tuberculosis Rv0792c, a GntR homolog in 25 mM HEPES Buffer; 400 mM NaCl, pH 7.2 were collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) using a Mythen 1K detector at a sample-detector distance of 0.3 m and at a wavelength of λ = 0.15414 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). One solute concentration of 3.20 mg/ml was measured at 10°C. Three successive 3600 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
Protein was freshly purified by gel filtration before data collection. SAXS data was collected using line collimation source. The three runs were desmeared and averaged using beam profile using SAXSQuant program. Final I(q) profile was analyzed using ATSAS programs. SAXS profile and parameters were used to steer missing lobes in the homology model to generate a composite model of the dimeric state of protein in solution. Data provided here shows: 1) Desmeared sample minus buffer file; 2) Damfilt and Damaver models; Its Average Fir file of the ten runs; and 3) CRYSOL program based comparison of computed SAXS profile of the atomistic model of protein w SAXS data
|
|
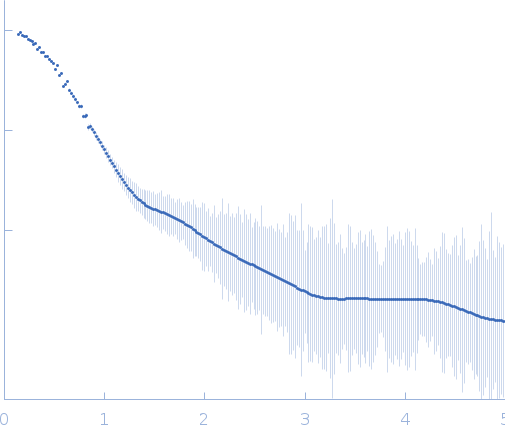 s, nm-1
s, nm-1