|
Synchrotron SAXS data from solutions of the relaxase domain of TraI in 50 mM TRIS-HCl, pH 8, 100 mM NaCl, were collected on the BM29 beam line at the ESRF storage ring (Grenoble, France) using a Pilatus 1M detector at a sample-detector distance of 2.9 m and at a wavelength of λ = 0.099 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Possible aggregates were removed using a Superdex 200 10/300 (GE Healthcare) size-exclusion chromatography (SEC) column pre-equilibrated with buffer at a flow rate of 0.5 mL/min at the EMBL-Lab outstation (Grenoble). All measurements were performed at 4°C with a sample injection concentration of 19.7 mg/mL. For online-SEC one frame was collected every two seconds and for static measurements ten frames of data were collected with total exposure time of one second. By comparison of these frames, the possibility of radiation damage during the measurement was excluded. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted. All used programs for data processing were part of the ATSAS Software package (Version 2.8.4).
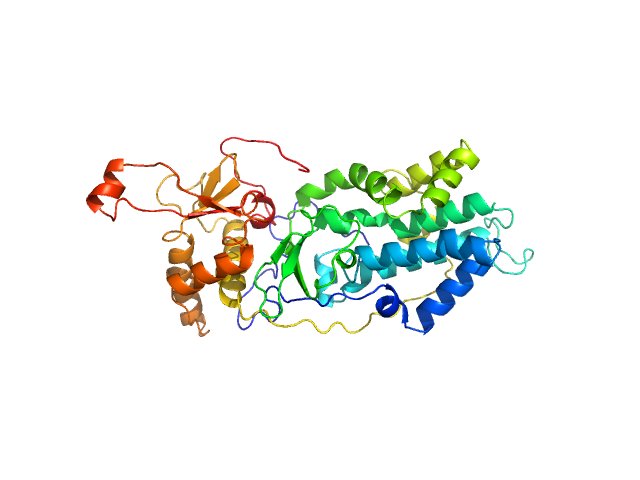
Please note: The protein contains unstructured regions that impart a level of flexibility to the overall structure in solution. As a consequence, the conformational state(s) sampled by the protein differs from - and cannot be captured by - the single predicted PHYRE2 atomistic model/representation displayed in this entry (as evidenced by the poor fit of the model to the SAXS data; bottom).
|
|
 s, nm-1
s, nm-1