|
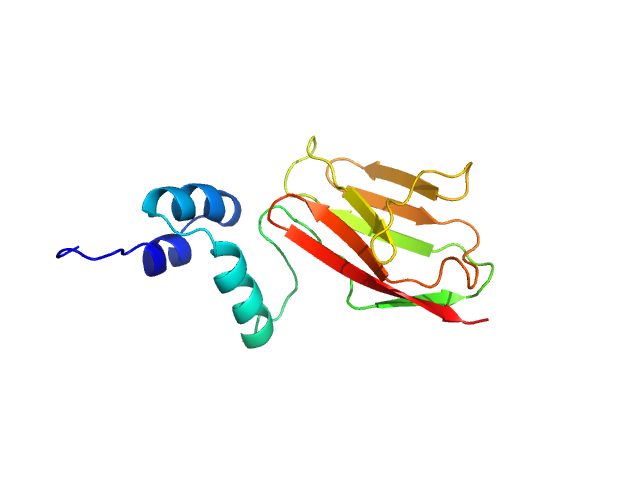
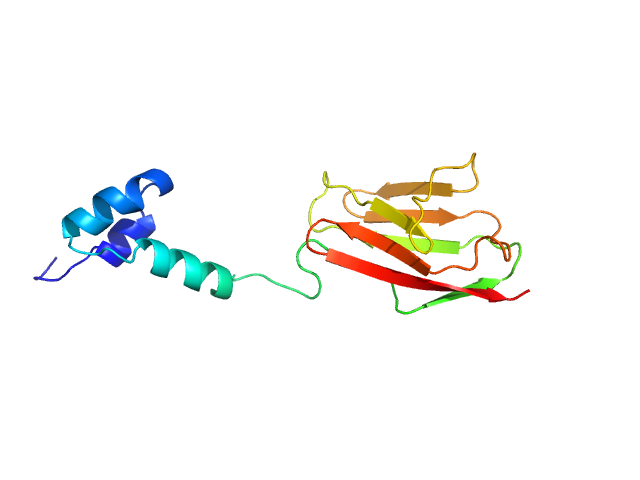
Synchrotron SAXS data from solutions of the cardiac myosin binding protein C tri-helixmotif bundle and C2 domain in 150 mM NaCl, 10 mM MES, 2 mM TCEP, 1 mM NaN3, pH 6.5 (at 4°C) were collected at the SAXS/WAXS beam line at the Australian Synchrotron (Melbourne, Australia) using a Pilatus 1M detector at a sample-detector distance of 2.7 m and at a wavelength of λ = 0.10332 nm (l(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). Solute concentrations ranging between 1.0 and 4.9 mg/ml were measured in batch mode. Data were reduced to I(s) versus s using the software ScatterBrain (http://www.synchrotron.org.au/aussyncbeamlines/saxswaxs/software-saxswaxs). Intensities were placed on an absolute scale using the known scattering from H2O. Samples were prepared by gel fitration with a solvent scattering blank taken from buffer eluted from the gel filtration column was measured before each sample. Scattering profiles were obtained by subtraction of the solvent blank scattering from the protein+solvent scattering and extrapolated to zero concentration in primus qt.
The concentration series SAXS data and EOM models are provided in the full entry zip archive.
|
|
 s, nm-1
s, nm-1
 Rg, nm
Rg, nm