|
|
|
|
|
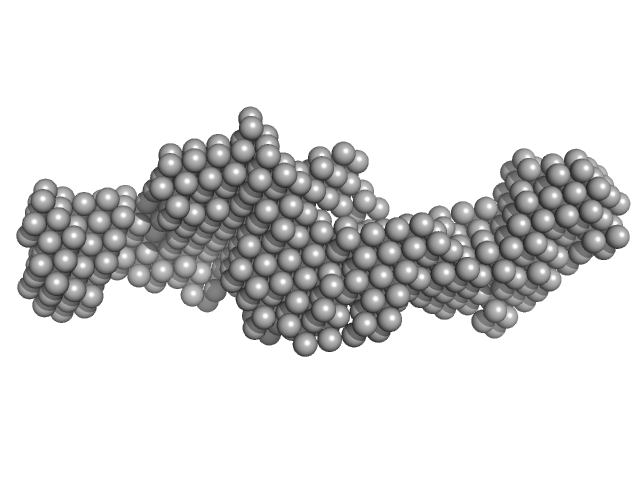
| Sample: |
Nuclear fragile X mental retardation-interacting protein 2 monomer, 59 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl pH 8.0, 5 mM TCEP, pH:
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 11
|
A study of the ultrastructure of fragile-X-related proteins.
Biochem J 419(2):347-57 (2009)
Sjekloća L, Konarev PV, Eccleston J, Taylor IA, Svergun DI, Pastore A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial trimer, 80 kDa Homo sapiens protein
|
| Buffer: |
20 mM citrate buffer, 1 mM DTT, 0.1 mM PMSF and 1 mM MgCl2, pH: 5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Feb 1
|
Molecular shape and prominent role of β-strand swapping in organization of dUTPase oligomers
FEBS Letters 583(5):865-871 (2009)
Takács E, Barabás O, Petoukhov M, Svergun D, Vértessy B
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Glutamate--tRNA ligase dimer, 108 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
35 mM HEPES⁄NaOH, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 19
|
Kinetic and mechanistic characterization of Mycobacterium tuberculosis glutamyl-tRNA synthetase and determination of its oligomeric structure in solution
FEBS Journal 276(5):1398-1417 (2009)
Paravisi S, Fumagalli G, Riva M, Morandi P, Morosi R, Konarev P, Petoukhov M, Bernier S, Chênevert R, Svergun D, Curti B, Vanoni M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
aD11 Fab dimer, 50 kDa Mus musculus protein
|
| Buffer: |
50 mM Na-phosphate 1mM ethylenediaminetetraacetic aci, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 20
|
Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J Mol Biol 381(4):881-96 (2008)
Covaceuszach S, Cassetta A, Konarev PV, Gonfloni S, Rudolph R, Svergun DI, Lamba D, Cattaneo A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
NGF dimer, 50 kDa Mus musculus protein
|
| Buffer: |
10 mM Na-phosphate 150 mM NaCl, pH: 7.8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 20
|
Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J Mol Biol 381(4):881-96 (2008)
Covaceuszach S, Cassetta A, Konarev PV, Gonfloni S, Rudolph R, Svergun DI, Lamba D, Cattaneo A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
aD11 Fab dimer, 50 kDa Mus musculus protein
NGF dimer, 50 kDa Mus musculus protein
|
| Buffer: |
30 mM Na-phosphate 75 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Dec 20
|
Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J Mol Biol 381(4):881-96 (2008)
Covaceuszach S, Cassetta A, Konarev PV, Gonfloni S, Rudolph R, Svergun DI, Lamba D, Cattaneo A
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Anaerobic nitric oxide reductase flavorubredoxin tetramer, 217 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 18% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 19
|
Quaternary Structure of Flavorubredoxin as Revealed by Synchrotron Radiation Small-Angle X-Ray Scattering
Structure 16(9):1428-1436 (2008)
Petoukhov M, Vicente J, Crowley P, Carrondo M, Teixeira M, Svergun D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Der p21, allegren dimer, 26 kDa Escherichia coli protein
|
| Buffer: |
10 mM Hepes , pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 24
|
Characterization of Der p 21, a new important allergen derived from the gut of house dust mites.
Allergy 63(6):758-67 (2008)
Weghofer M, Dall'Antonia Y, Grote M, Stöcklinger A, Kneidinger M, Balic N, Krauth MT, Fernández-Caldas E, Thomas WR, van Hage M, Vieths S, Spitzauer S, Horak F, Svergun DI, Konarev PV, Valent P, Thalh...
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytochrome C monomer, 11 kDa Escherichia coli protein
Adrenodoxin monomer, 11 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES 2 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jun 15
|
Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy.
J Am Chem Soc 130(20):6395-403 (2008)
Xu X, Reinle W, Hannemann F, Konarev PV, Svergun DI, Bernhardt R, Ubbink M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytochrome C dimer dimer, 22 kDa Escherichia coli protein
Adrenodoxin dimer dimer, 22 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES 2 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jun 15
|
Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy.
J Am Chem Soc 130(20):6395-403 (2008)
Xu X, Reinle W, Hannemann F, Konarev PV, Svergun DI, Bernhardt R, Ubbink M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
64 |
nm3 |
|
|