|
|
|
|

|
| Sample: |
pfyP - Blue light photoreceptor dimer, 58 kDa Bacillus subtilis protein
|
| Buffer: |
PBS + 5 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 24
|
The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS stu...
J Mol Biol 403(1):78-87 (2010)
Jurk M, Dorn M, Kikhney A, Svergun D, Gärtner W, Schmieder P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
pfyP - Blue light photoreceptor dimer, 58 kDa Bacillus subtilis protein
|
| Buffer: |
PBS + 5 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 24
|
The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS stu...
J Mol Biol 403(1):78-87 (2010)
Jurk M, Dorn M, Kikhney A, Svergun D, Gärtner W, Schmieder P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Membrane scaffold protein 1D1 dimer, 50 kDa unidentified protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 100 mM sodium cholate,, pH: 7.4
|
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2009 Nov 14
|
Elliptical structure of phospholipid bilayer nanodiscs encapsulated by scaffold proteins: casting the roles of the lipids and the protein.
J Am Chem Soc 132(39):13713-22 (2010)
Skar-Gislinge N, Simonsen JB, Mortensen K, Feidenhans'l R, Sligar SG, Lindberg Møller B, Bjørnholm T, Arleth L
|
| RgGuinier |
4.9 |
nm |
| Dmax |
12.2 |
nm |
|
|
|
|
|
|

|
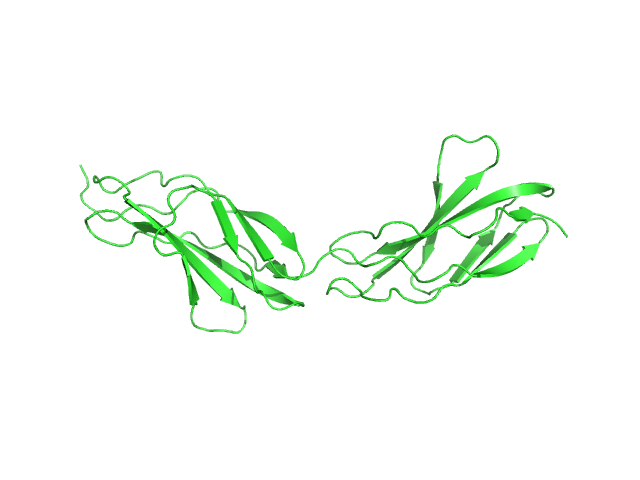
| Sample: |
Phosphoenolpyruvate-protein phosphotransferase dimer, 127 kDa Escherichia coli protein
|
| Buffer: |
20mM TRIS buffer, 100 mM NaCl, 10 mM DTT, 4 mM MgCl2, 1 mM EDTA, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12-ID-C, Advanced Photon Source (APS), Argonne National Laboratory on 2010 Aug 23
|
Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering.
J Am Chem Soc 132(37):13026-45 (2010)
Schwieters CD, Suh JY, Grishaev A, Ghirlando R, Takayama Y, Clore GM
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
|
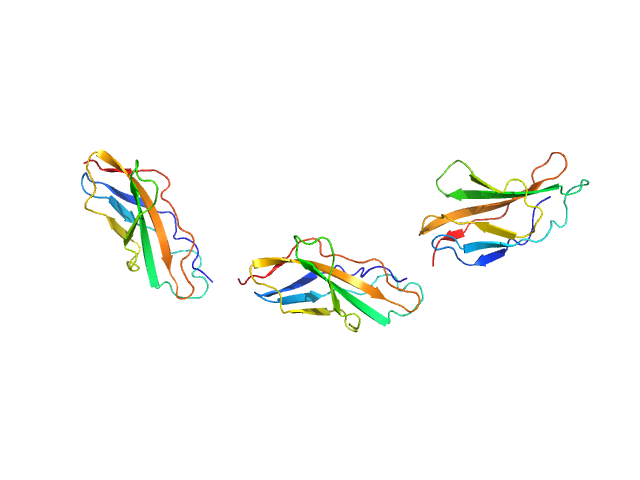
| Sample: |
Titin monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 3
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
2.5 |
nm |
| Dmax |
90.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
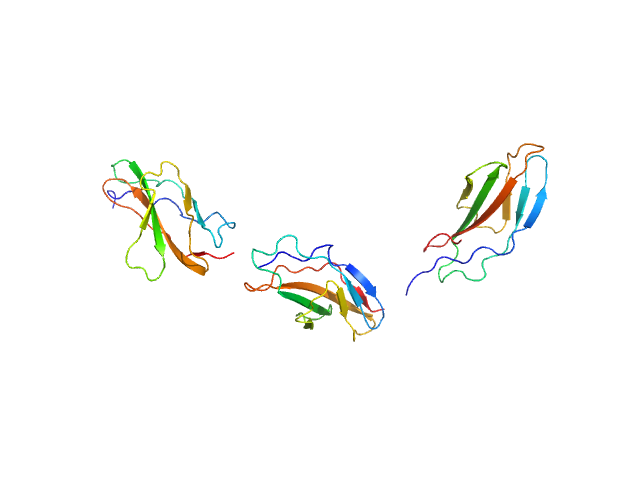
| Sample: |
Titin monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 3
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
3.7 |
nm |
| Dmax |
130.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Titin monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 6
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Factor H CCP modules 10 to 15 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Potassium Phosphate, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Dec 2
|
The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module.
J Mol Biol 395(1):105-22 (2010)
Schmidt CQ, Herbert AP, Mertens HD, Guariento M, Soares DC, Uhrin D, Rowe AJ, Svergun DI, Barlow PN
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Factor H CCP modules 11 to 14 monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
50 mM Potassium Phosphate, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Dec 2
|
The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module.
J Mol Biol 395(1):105-22 (2010)
Schmidt CQ, Herbert AP, Mertens HD, Guariento M, Soares DC, Uhrin D, Rowe AJ, Svergun DI, Barlow PN
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Factor H CCP modules 12 to 13 monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
50 mM Potassium Phosphate, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Dec 2
|
The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module.
J Mol Biol 395(1):105-22 (2010)
Schmidt CQ, Herbert AP, Mertens HD, Guariento M, Soares DC, Uhrin D, Rowe AJ, Svergun DI, Barlow PN
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
20 |
nm3 |
|
|