|
|
|
|

|
| Sample: |
Pyruvate decarboxylase tetramer, 244 kDa Zymomonas mobilis protein
|
| Buffer: |
100 mM Sodium Citrate, 17% Glycerol, 22.5% PEG 1500, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 1998 Nov 3
|
Crystal versus solution structures of thiamine diphosphate-dependent enzymes.
J Biol Chem 275(1):297-302 (2000)
Svergun DI, Petoukhov MV, Koch MH, König S
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
264 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.3 |
nm |
| Dmax |
18.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.5 |
nm |
|
|
|
|
|
|
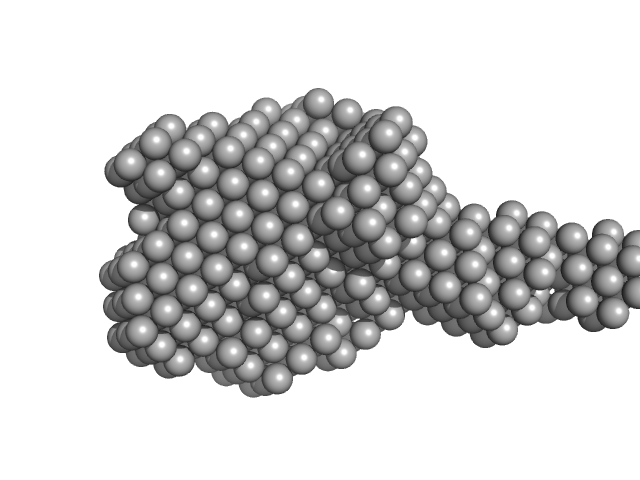
|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
231 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Structural polyprotein dimer, 26 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 27
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Structural polyprotein (Capsid protein VP3: K947R; Δ756-843; Δ977-1012) monomer, 18 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM TRIS, 500 mM NaCl, 2 mM DTT,, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Structural polyprotein (Capsid protein VP3: K947R; Δ977-1012) dimer, 55 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM TRIS, 500 mM NaCl, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 1
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|