|
|
|
|
|
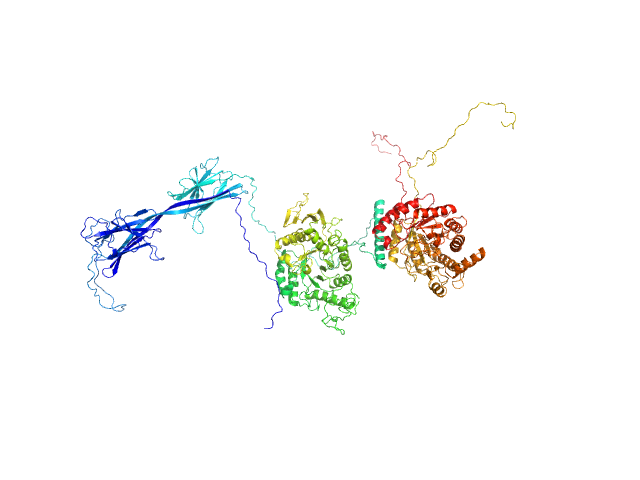
| Sample: |
Inactive beta-amylase 9 monomer, 50 kDa Arabidopsis thaliana protein
Alpha-amylase 3, chloroplastic monomer, 94 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 0.2 mM TCEP, pH: 7
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 May 16
|
The Pseudoenzyme β‐Amylase9 From Arabidopsis Activates α‐Amylase3: A Possible Mechanism to Promote Stress‐Induced Starch Degradation
Proteins: Structure, Function, and Bioinformatics (2025)
Berndsen C, Storm A, Sardelli A, Hossain S, Clermont K, McFather L, Connor M, Monroe J
|
| RgGuinier |
5.0 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
380 |
nm3 |
|