|
|
|
|
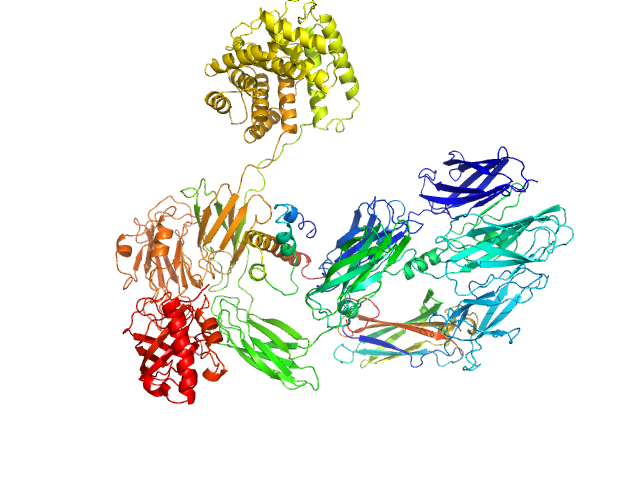
|
| Sample: |
Complement C3 (Δ668-671) monomer, 187 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES pH 6.0, 200 mM NaCl, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 7
|
Cryo-EM analysis of complement C3 reveals a reversible major opening of the macroglobulin ring.
Nat Struct Mol Biol (2025)
...Jørgensen MH, Olesen HG, Lorentzen J, Harwood SL, Almeida AV, Fruergaard MU, Jensen RK, Kanis P, Pedersen H, Tranchant E, Petersen SV, Thøgersen IB, Kragelund BB, Lyons JA, Enghild JJ, Andersen GR
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
357 |
nm3 |
|