|
|
|
|
|
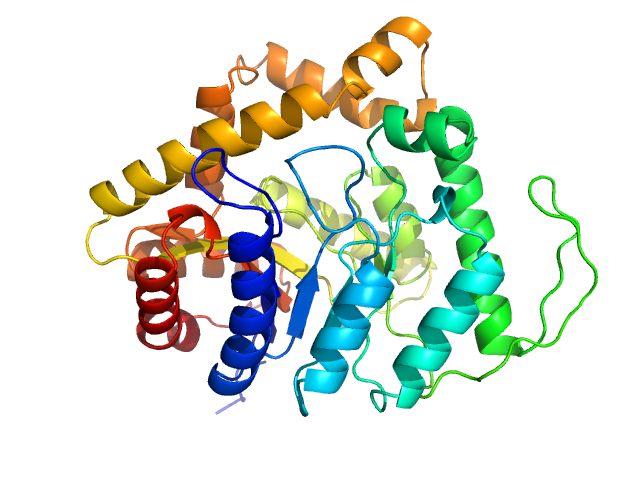
| Sample: |
Alkanal monooxygenase alpha chain dimer, 84 kDa Enhygromyxa salina protein
|
| Buffer: |
150 mM NaCl, 25 mM Tris, pH: 8
|
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2020 Aug 1
|
luxA
Gene From Enhygromyxa salina
Encodes a Functional Homodimeric Luciferase
Proteins: Structure, Function, and Bioinformatics (2024)
...Sluchanko N, Ryzhykau Y, Semenov Y, Kuklin A, Manukhov I, Gushchin I
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
138 |
nm3 |
|