|
|
|
|
|
| Sample: |
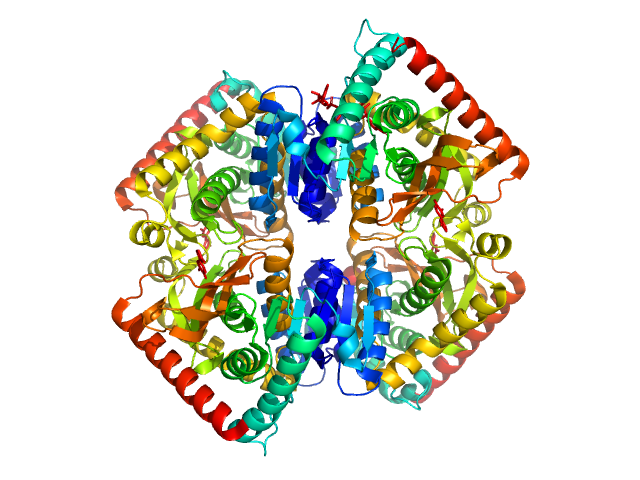
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology 4(1) (2021)
...Taube M, Kozak M, Holak T, Dömling A, Groves M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
211 |
nm3 |
|