|
|
|
|
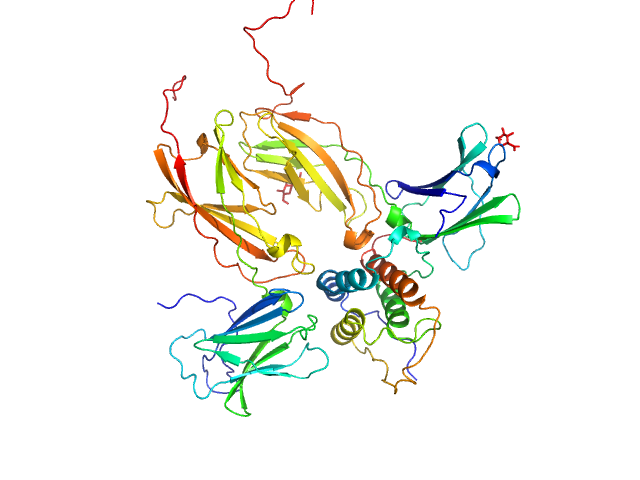
|
| Sample: |
Thymic stromal lymphopoietin monomer, 15 kDa Homo sapiens protein
Interleukin-7 receptor subunit alpha monomer, 26 kDa Homo sapiens protein
Cytokine receptor-like factor 2 monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and antagonism of the receptor complex mediated by human TSLP in allergy and asthma.
Nat Commun 8:14937 (2017)
Verstraete K, Peelman F, Braun H, Lopez J, Van Rompaey D, Dansercoer A, Vandenberghe I, Pauwels K, Tavernier J, Lambrecht BN, Hammad H, De Winter H, Beyaert R, Lippens G, Savvides SN
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
98 |
nm3 |
|