| MWI(0) | 39 | kDa |
| MWexpected | 38 | kDa |
| VPorod | 54 | nm3 |
|
log I(s)
2.76×10-2
2.76×10-3
2.76×10-4
2.76×10-5
|
 s, nm-1
s, nm-1
|
|
|
|

|
|
|
|
|
|
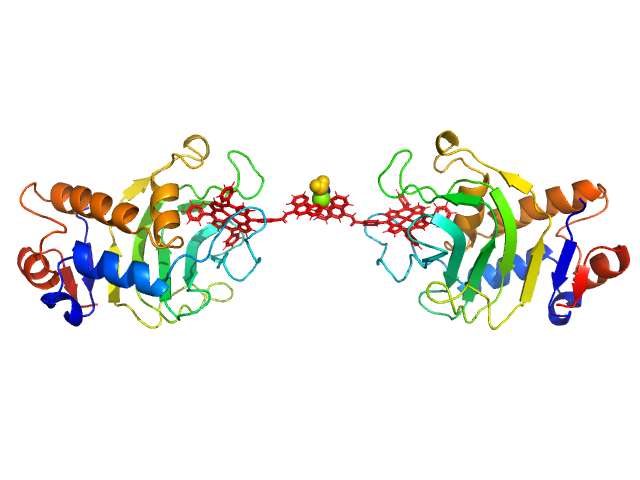
|
|
|
|
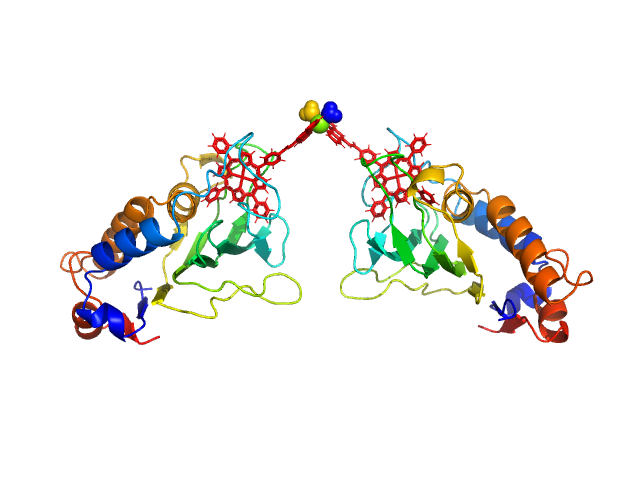
|
|
|
|
Synchrotron SAXS data from solutions of heme acquisition system protein A in 50 mM CHES, 5 % glycerol, pH 9.5 were collected on the BL-10C beam line at the Photon Factory (PF), High Energy Accelerator Research Organization (KEK; Tsukuba, Japan) using a Pilatus3 2M detector at a sample-detector distance of 2.1 m and at a wavelength of λ = 0.1 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). In-line size-exclusion chromatography (SEC) SAS was employed. The SEC parameters were as follows: A 440.00 μl sample at 8 mg/ml was injected at a 0.05 ml/min flow rate onto a Cytiva Superdex 200 Increase 10/300 column at 20°C. 408 successive 20 second frames were collected. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
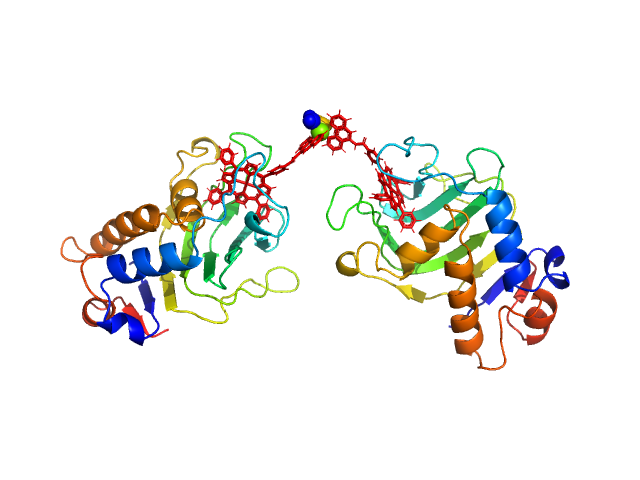
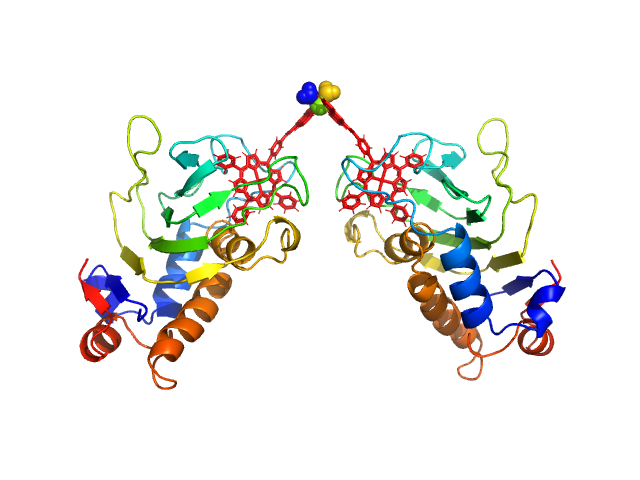
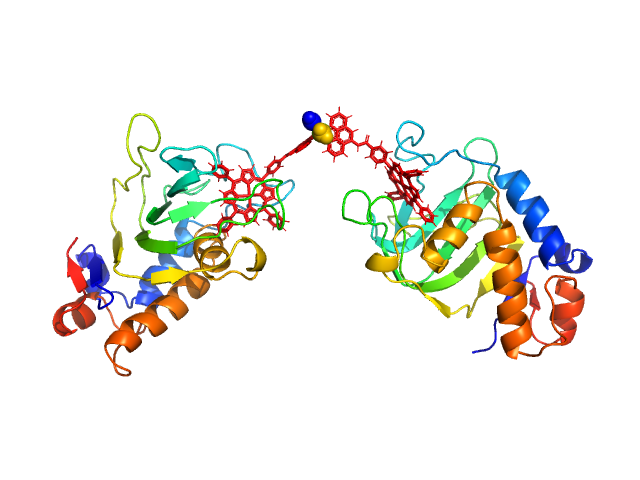
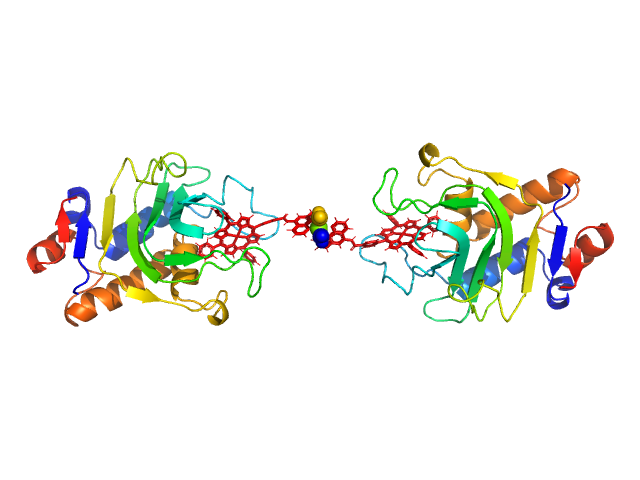
HasApf5 incorporating iron(III)-tetraphenylporphyrin (TPP) with the phenyl ligand attached to the phenyl group of Fe-TPP via a planar amide bond (Fe-TPP-phen) formed via a dimer of Ni2+ ions. The molecular weight of the dimer containing Fe-TPP-phen was estimated to be 39.9 kDa. This SAXS profile was compared with six atomic model structures (delta-cis.pdb, delta-trans1.pdb, delta-trans2.pdb, lambda-cis.pdb, lambda-trans1.pdb, lambda-trans2.pdb) that were created based on crystal structures. The methods to create these model structures are described in detail in the Supplementary Information of the article. Since lambda-trans1.pdb was shown to be the closest to the experimental results on the basis of the CRYSOL calculations, the remaining five models were superimposed on lambda-trans1.pdb using the software BIOVIA Discovery Studio. |
|
|||||||||||||||||||||||||||