|
|
|
|

|
| Sample: |
N-(2-hydroxypropyl)- 31 methacrylamide (HPMA) copolymers with Cholesterol (2.7%) 0, 16740 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 7.2), pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 27
|
Macromolecular HPMA-based nanoparticles with cholesterol for solid-tumor targeting: detailed study of the inner structure of a highly efficient drug delivery system.
Biomacromolecules 13(8):2594-604 (2012)
Filippov SK, Chytil P, Konarev PV, Dyakonova M, Papadakis C, Zhigunov A, Plestil J, Stepanek P, Etrych T, Ulbrich K, Svergun DI
|
| RgGuinier |
5.2 |
nm |
| Dmax |
28.1 |
nm |
|
|
|
|
|
|

|
| Sample: |
N-(2-hydroxypropyl)- 31 methacrylamide (HPMA) copolymers with Cholesterol (3.0%) 0, 29520 kDa
|
| Buffer: |
phosphate buffer saline (PBS) (pH 5.0), pH: 5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 27
|
Macromolecular HPMA-based nanoparticles with cholesterol for solid-tumor targeting: detailed study of the inner structure of a highly efficient drug delivery system.
Biomacromolecules 13(8):2594-604 (2012)
Filippov SK, Chytil P, Konarev PV, Dyakonova M, Papadakis C, Zhigunov A, Plestil J, Stepanek P, Etrych T, Ulbrich K, Svergun DI
|
| RgGuinier |
9.4 |
nm |
| Dmax |
43.2 |
nm |
|
|
|
|
|
|
|
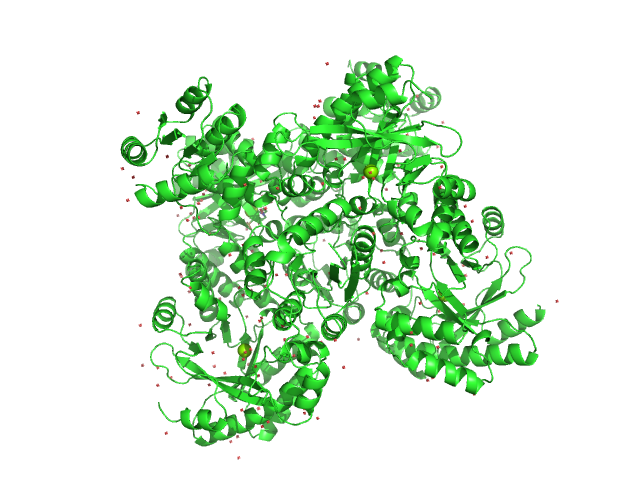
| Sample: |
RocR tetramer, 171 kDa Pseudomonas aeruginosa (strain … protein
|
| Buffer: |
50 mM Tris–HCl, 250 mM NaCl, 10 mM imidazole, 5% glycerol, 0.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 9
|
Structural Insights into the Regulatory Mechanism of the Response Regulator RocR from Pseudomonas aeruginosa in Cyclic Di-GMP Signaling
Journal of Bacteriology 194(18):4837-4846 (2012)
Chen M, Kotaka M, Vonrhein C, Bricogne G, Rao F, Chuah M, Svergun D, Schneider G, Liang Z, Lescar J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
|
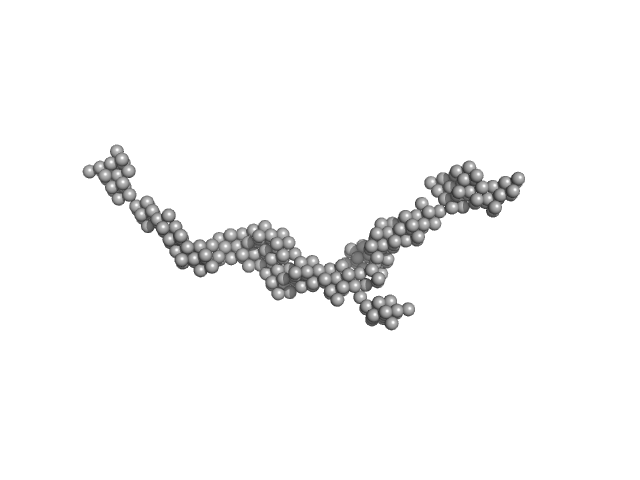
| Sample: |
Myomesin-1 dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
25 mMTris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 10
|
Superhelical architecture of the myosin filament-linking protein myomesin with unusual elastic properties.
PLoS Biol 10(2):e1001261 (2012)
Pinotsis N, Chatziefthimiou SD, Berkemeier F, Beuron F, Mavridis IM, Konarev PV, Svergun DI, Morris E, Rief M, Wilmanns M
|
| RgGuinier |
9.6 |
nm |
| Dmax |
37.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Plectin monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 2.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2009 Jul 24
|
The structure of the plakin domain of plectin reveals a non-canonical SH3 domain interacting with its fourth spectrin repeat.
J Biol Chem 286(14):12429-38 (2011)
Ortega E, Buey RM, Sonnenberg A, de Pereda JM
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Multidomain regulatory protein Rv1364c monomer, 70 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 14
|
Structural characterization of the multidomain regulatory protein Rv1364c from Mycobacterium tuberculosis.
Structure 19(1):56-69 (2011)
King-Scott J, Konarev PV, Panjikar S, Jordanova R, Svergun DI, Tucker PA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Multidomain regulatory protein Rv1364c dimer, 139 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 14
|
Structural characterization of the multidomain regulatory protein Rv1364c from Mycobacterium tuberculosis.
Structure 19(1):56-69 (2011)
King-Scott J, Konarev PV, Panjikar S, Jordanova R, Svergun DI, Tucker PA
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
289 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Protein A monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
10 mM HEPES, pH 7.2, 3 mM EDTA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 12
|
A structural basis for Staphylococcal complement subversion: X-ray structure of the complement-binding domain of Staphylococcus aureus protein Sbi in complex with ligand C3d
Molecular Immunology 48(4):452-462 (2011)
Clark E, Crennell S, Upadhyay A, Zozulya A, Mackay J, Svergun D, Bagby S, van den Elsen J
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PfyP - Blue light photoreceptor dimer, 58 kDa Bacillus subtilis protein
|
| Buffer: |
PBS + 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Oct 24
|
The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS study.
J Mol Biol 403(1):78-87 (2010)
Jurk M, Dorn M, Kikhney A, Svergun D, Gärtner W, Schmieder P
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
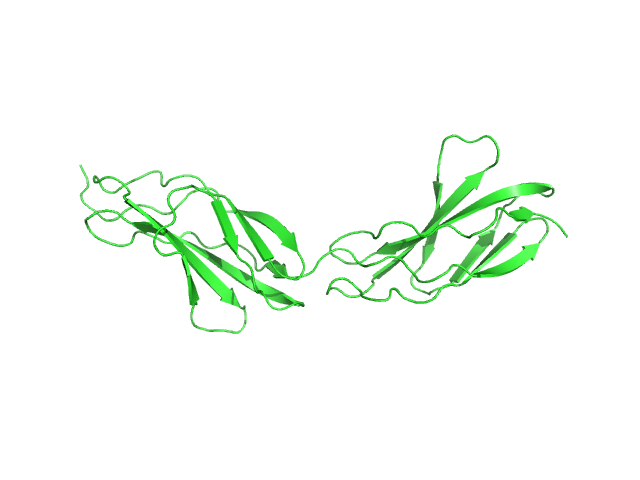
| Sample: |
Titin monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Jul 3
|
The Structure of the FnIII Tandem A77-A78 Points to a Periodically Conserved Architecture in the Myosin-Binding Region of Titin
Journal of Molecular Biology 401(5):843-853 (2010)
Bucher R, Svergun D, Muhle-Goll C, Mayans O
|
| RgGuinier |
2.5 |
nm |
| Dmax |
90.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|