|
|
|
|

|
| Sample: |
Mouse ProNGF - pro-form of the NGF (nerve growth factor) dimer, 100 kDa Mus musculus protein
|
| Buffer: |
50 mM Na-phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 18
|
Structural and functional properties of mouse proNGF.
Biochem Soc Trans 34(Pt 4):605-6 (2006)
Paoletti F, Konarev PV, Covaceuszach S, Schwarz E, Cattaneo A, Lamba D, Svergun DI
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polypyrimidine tract-binding protein 2 monomer, 57 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 250 mM NaCl, 2 mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 11
|
Structure and RNA interactions of the N-terminal RRM domains of PTB.
Structure 12(9):1631-43 (2004)
Simpson PJ, Monie TP, Szendröi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthews S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Procollagen C-endopeptidase enhancer 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Jul 9
|
Low Resolution Structure Determination Shows Procollagen C-Proteinase Enhancer to be an Elongated Multidomain Glycoprotein
Journal of Biological Chemistry 278(9):7199-7205 (2003)
Bernocco S, Steiglitz B, Svergun D, Petoukhov M, Ruggiero F, Ricard-Blum S, Ebel C, Geourjon C, Deléage G, Font B, Eichenberger D, Greenspan D, Hulmes D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
264 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.3 |
nm |
| Dmax |
18.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.5 |
nm |
|
|
|
|
|
|
|
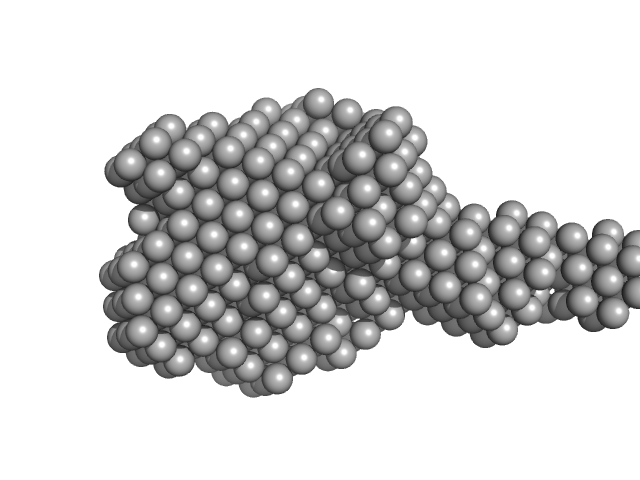
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
231 |
nm3 |
|
|