|
|
|
|

|
| Sample: |
Tyrosine-protein kinase BTK (R28C mutant) monomer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl, 2 mM DTT and 1 mM MgCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Apr 2
|
Conformation of full-length Bruton tyrosine kinase (Btk) from synchrotron X-ray solution scattering.
EMBO J 22(18):4616-24 (2003)
Márquez JA, Smith CI, Petoukhov MV, Lo Surdo P, Mattsson PT, Knekt M, Westlund A, Scheffzek K, Saraste M, Svergun DI
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ferredoxin-dependent glutamate synthase 2 monomer, 169 kDa Synechocystis sp. (strain … protein
Ferredoxin-1 monomer, 11 kDa Nostoc sp. (strain … protein
|
| Buffer: |
Hepes– KOH buffer, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Jun 4
|
The Active Conformation of Glutamate Synthase and its Binding to Ferredoxin
Journal of Molecular Biology 330(1):113-128 (2003)
van den Heuvel R, Svergun D, Petoukhov M, Coda A, Curti B, Ravasio S, Vanoni M, Mattevi A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Procollagen C-endopeptidase enhancer 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Jul 9
|
Low Resolution Structure Determination Shows Procollagen C-Proteinase Enhancer to be an Elongated Multidomain Glycoprotein
Journal of Biological Chemistry 278(9):7199-7205 (2003)
Bernocco S, Steiglitz B, Svergun D, Petoukhov M, Ruggiero F, Ricard-Blum S, Ebel C, Geourjon C, Deléage G, Font B, Eichenberger D, Greenspan D, Hulmes D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Pyruvate decarboxylase tetramer, 244 kDa Zymomonas mobilis protein
|
| Buffer: |
100 mM Sodium Citrate, 17% Glycerol, 22.5% PEG 1500, pH: 6
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 1998 Nov 3
|
Crystal versus solution structures of thiamine diphosphate-dependent enzymes.
J Biol Chem 275(1):297-302 (2000)
Svergun DI, Petoukhov MV, Koch MH, König S
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
264 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE1 dimer, 179 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.3 |
nm |
| Dmax |
18.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.5 |
nm |
|
|
|
|
|
|
|
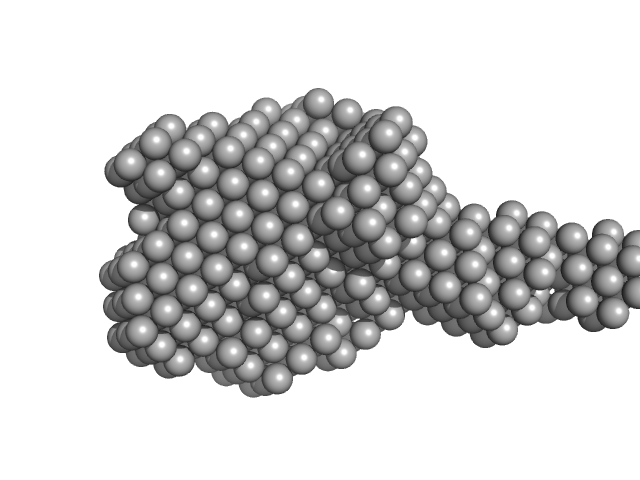
| Sample: |
Xylosyl- and glucuronyltransferase LARGE2 dimer, 168 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Mar 18
|
LARGE1 Processively Polymerizes Matriglycan Using Active Sites on Alternate Protomers
SSRN Electronic Journal ()
Joseph S, Schnicker N, Xu Z, Yang T, Hopkins J, Watkins M, Chakravarthy S, Davulcu O, Anderson M, Venzke D, Campbell K
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
231 |
nm3 |
|
|