|
|
|
|
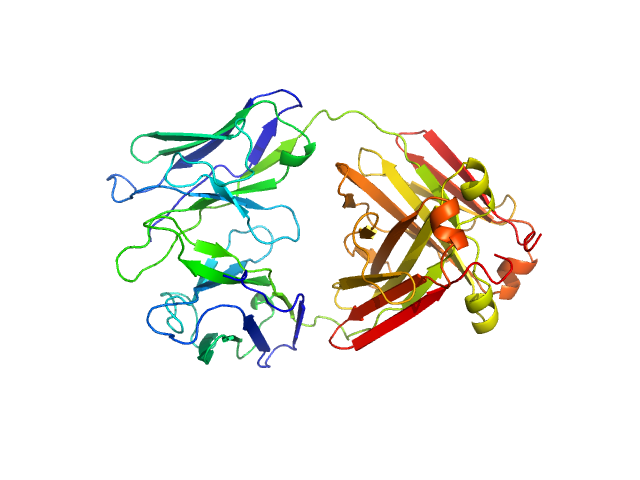
|
| Sample: |
Immunoglobulin light chain H3 dimer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 12
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
56 |
nm3 |
|