|
|
|
|
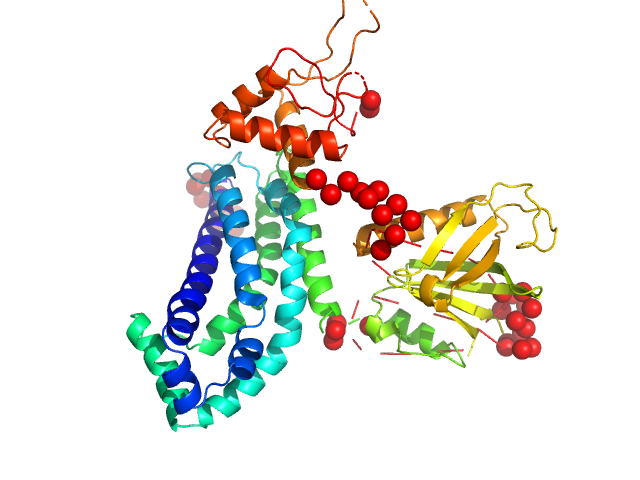
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7, 300 mM NaCl, 2% glycerol, 2 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 8
|
Structural and dynamic changes in P-Rex1 upon activation by PIP3 and inhibition by IP4
eLife 12 (2024)
Ravala S, Adame-Garcia S, Li S, Chen C, Cianfrocco M, Silvio Gutkind J, Cash J, Tesmer J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
79 |
nm3 |
|