|
|
|
|
|
| Sample: |
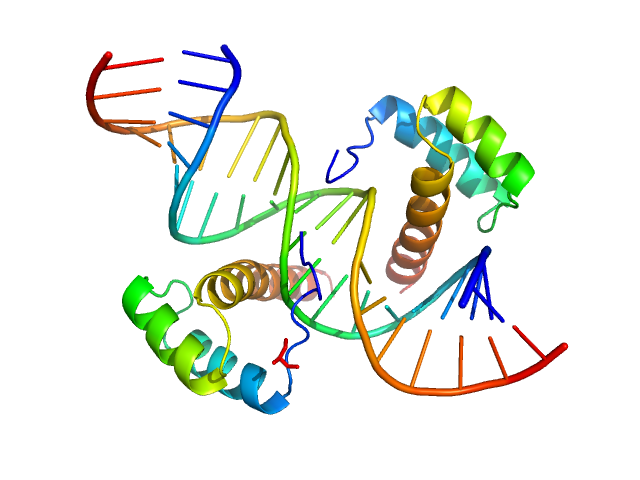
Cone-rod homeobox protein , 20 kDa Homo sapiens protein
Ret4 sense strand , 6 kDa synthetic construct DNA
Ret4 antisense strand , 6 kDa synthetic construct DNA
|
| Buffer: |
20 mM Tris, 150 mM KCl, 5 % glycerol, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 25
|
Molecular basis of CRX/DNA recognition and stoichiometry at the Ret4 response element
Structure (2024)
Srivastava D, Gowribidanur-Chinnaswamy P, Gaur P, Spies M, Swaroop A, Artemyev N
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
37 |
nm3 |
|