|
|
|
|
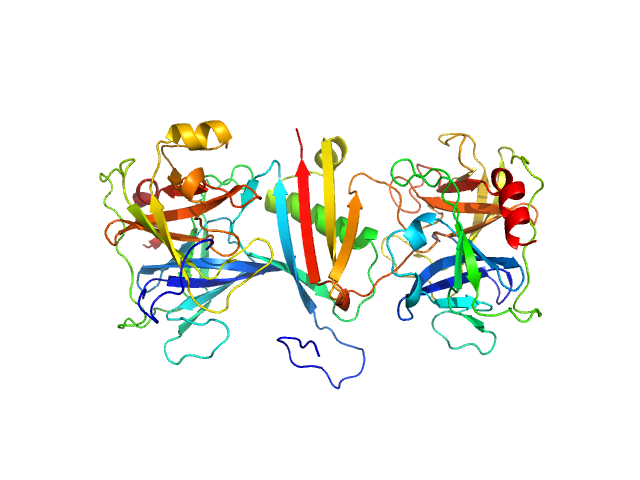
|
| Sample: |
Cathepsin G monomer, 25 kDa Homo sapiens protein
MAP domain-containing protein monomer, 13 kDa Staphylococcus aureus (strain … protein
Neutrophil elastase monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
77 |
nm3 |
|