|
|
|
|
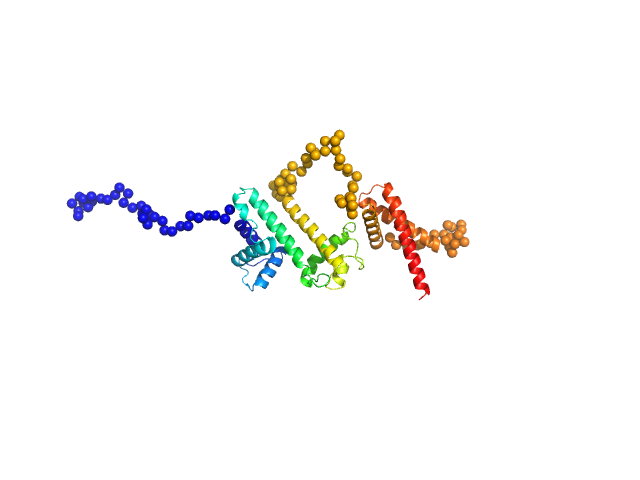
|
| Sample: |
Harpin Z2 monomer, 37 kDa Pseudomonas syringae strain … protein
|
| Buffer: |
10 mM MES, 100 mM NaF, pH: 6.2
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Centre for Cellular and Molecular Biology on 2023 May 1
|
Solution conformation of HrpZ2 protein from Pseudomonas syringae
Arpita Goswami
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
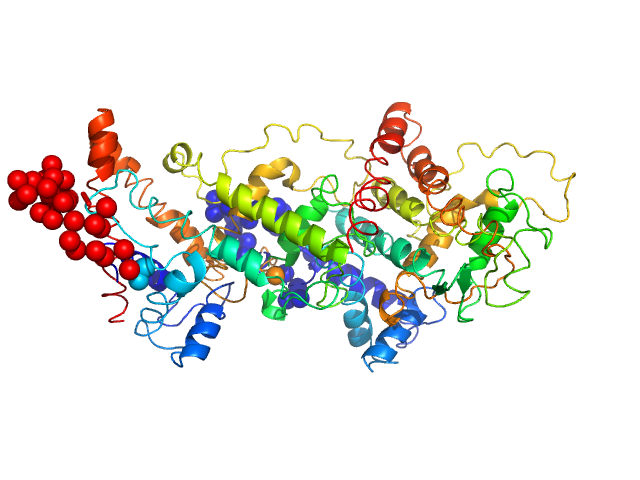
|
| Sample: |
Harpin Z2 monomer, 37 kDa Pseudomonas syringae strain … protein
|
| Buffer: |
10 mM MES, 100 mM NaF, pH: 6.2
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Centre for Cellular and Molecular Biology on 2023 May 1
|
Solution conformation of HrpZ2 protein from Pseudomonas syringae
Arpita Goswami
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
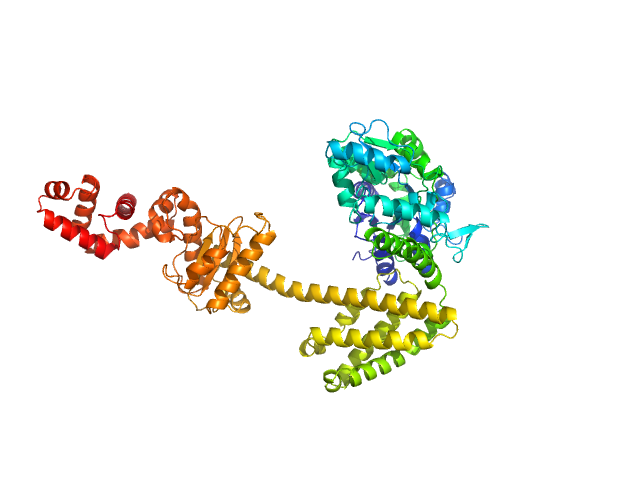
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) monomer, 40 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 monomer, 43 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH:
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
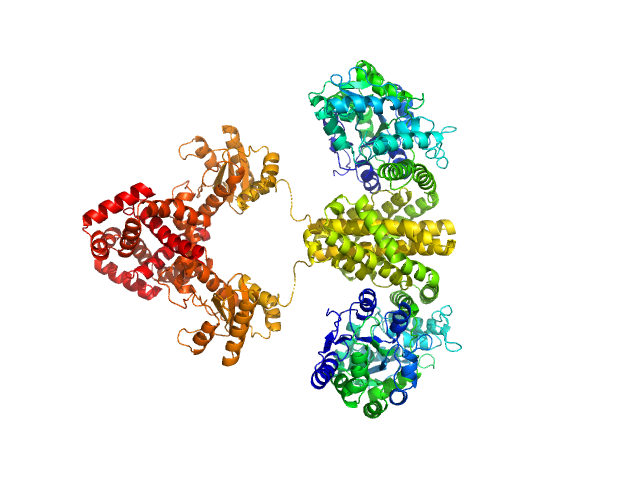
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) dimer, 80 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 dimer, 86 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH:
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
345 |
nm3 |
|
|
|
|
|
|
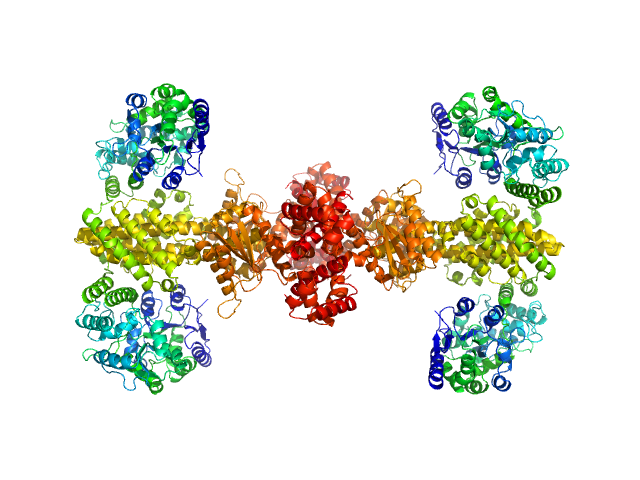
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) tetramer, 161 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 tetramer, 171 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH:
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
826 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ras-related protein Rab-1A monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 17
|
The SAXS Analysis of the human GTPase Rab1a
Wenhua Zhang
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphocholine hydrolase Lem3 monomer, 63 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Sep 20
|
The structural analysis of dephosphocholinase Legionella pneumophila Lem3
Wenhua Zhang
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Uncharacterized protein SAUSA300_1119 dimer, 125 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM KCl, 1 mM TCEP, 5% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 5
|
Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase.
J Biol Chem 300(12):107920 (2024)
Myers MJ, Xu Z, Ryan BJ, DeMars ZR, Ridder MJ, Johnson DK, Krute CN, Flynn TS, Kashipathy MM, Battaile KP, Schnicker N, Lovell S, Freudenthal BD, Bose JL
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
|
|
|
|
|
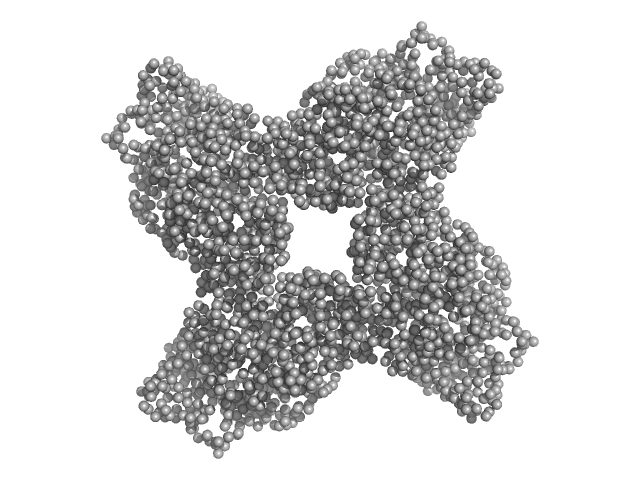
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
439 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
435 |
nm3 |
|
|