|
|
|
|

|
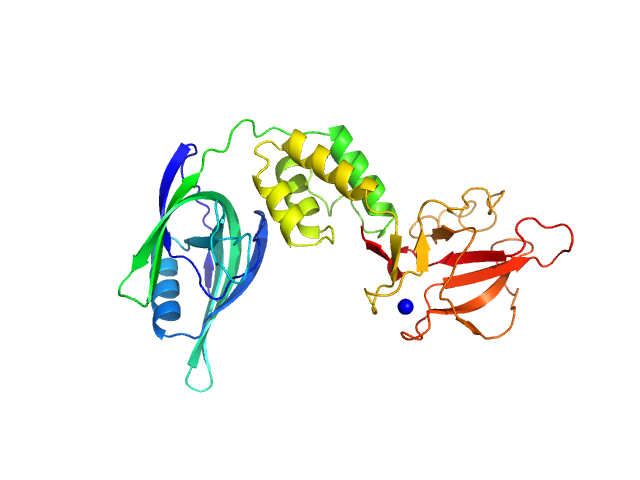
| Sample: |
Putative acylaminoacyl-peptidase tetramer, 293 kDa Bacillus spizizenii (strain … protein
|
| Buffer: |
10 mM Tris-HCl, 135 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2024 Mar 18
|
Structural adaptations for carboxypeptidase activity in putative S9 acylaminoacyl peptidase from Bacillus subtilis.
Int J Biol Macromol :136734 (2024)
Chandravanshi K, Singh R, Kumar A, Bhange GN, Kumar A, Makde RD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
446 |
nm3 |
|
|
|
|
|
|
|
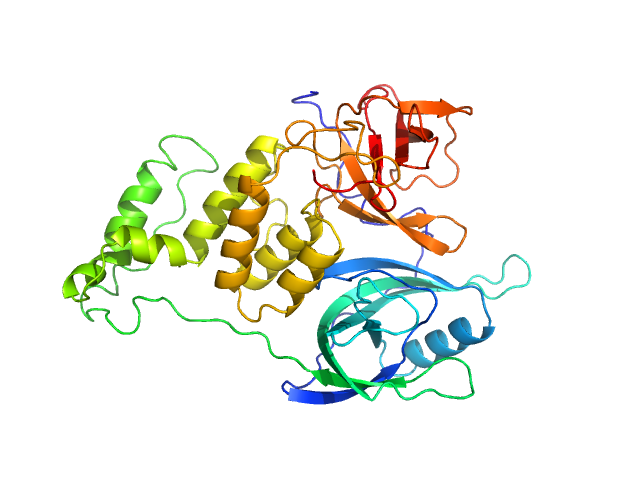
| Sample: |
Cereblon-midi monomer, 37 kDa protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D...
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
mezigdomide monomer, 1 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D...
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
pomalidomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D...
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Iberdomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D...
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Lenalidomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D...
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa synthetic construct protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 29
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D...
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa synthetic construct protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 29
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D...
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
G-quadrupex monomer, 6 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 15
|
G-quadruplex from HBV genome
Trushar Patel
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
G-quadruplex mutant monomer, 6 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jan 15
|
G-quadruplex from HBV genome
Trushar Patel
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
11 |
nm3 |
|
|