|
|
|
|
|
| Sample: |
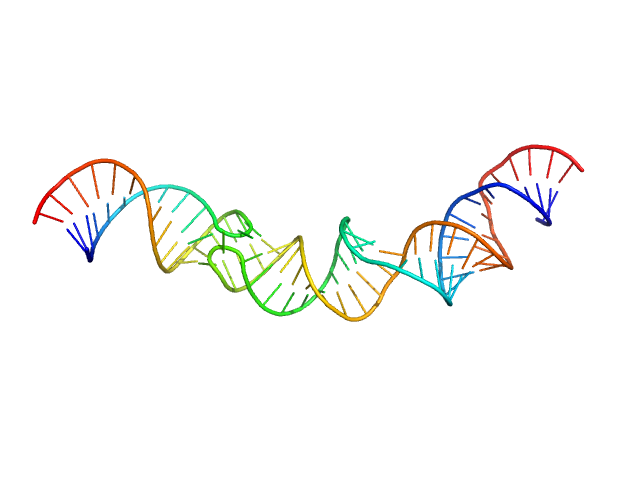
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
...Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
40 |
nm3 |
|