|
|
|
|
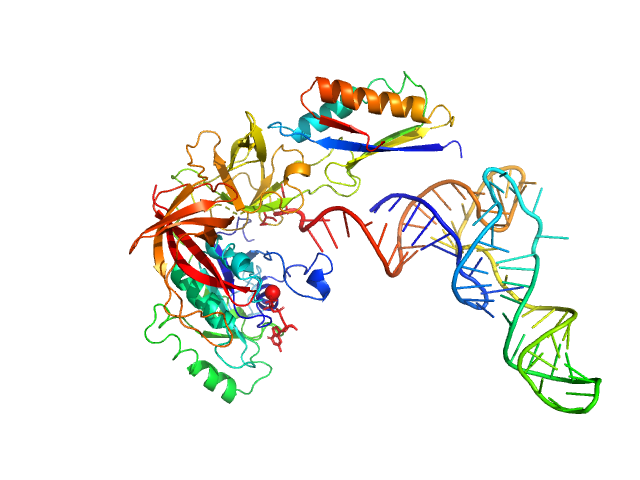
|
| Sample: |
Translation initiation factor 2 subunit gamma monomer, 46 kDa Saccharolobus solfataricus (strain … protein
Translation initiation factor 2 subunit alpha monomer, 10 kDa Saccharolobus solfataricus (strain … protein
transfer RNA monomer, 23 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS- NaOH pH 6.7, 200 mM NaCl, 5 mM MgCl 2, 1 mM GDPNP, pH: 6.7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
...Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|