|
|
|
|
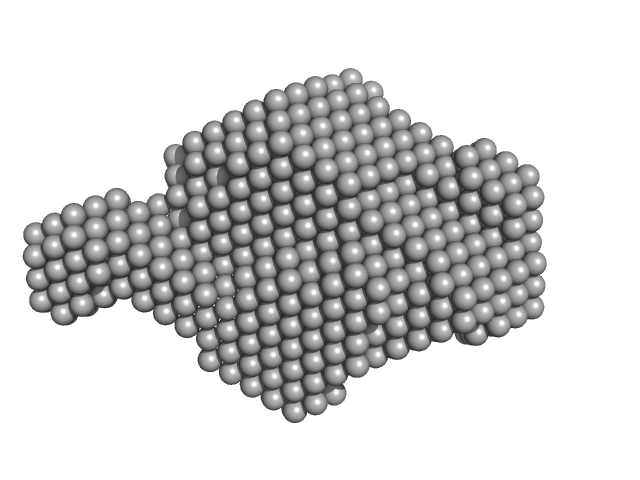
|
| Sample: |
Aureochrome 1a (N-terminally truncated) dimer, 53 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
20 mM HEPES 100 mM NaCl 10 mM MgCl2 5% w/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2015 Mar 11
|
Blue light-induced LOV domain dimerization enhances the affinity of Aureochrome 1a for its target DNA sequence.
Elife 5:e11860 (2016)
Heintz U, Schlichting I
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a-A´α-LOV-Jα dimer, 36 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 16
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a-A´α-LOV-Jα dimer, 36 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Mar 16
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a- A´α-LOV-Jα (Dark State) dimer, 32 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 9
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome1a- A´α-LOV-Jα (Light State) dimer, 32 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Sep 9
|
Structure of a Native-like Aureochrome 1a LOV Domain Dimer from Phaeodactylum tricornutum.
Structure 24(1):171-178 (2016)
Banerjee A, Herman E, Kottke T, Essen LO
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
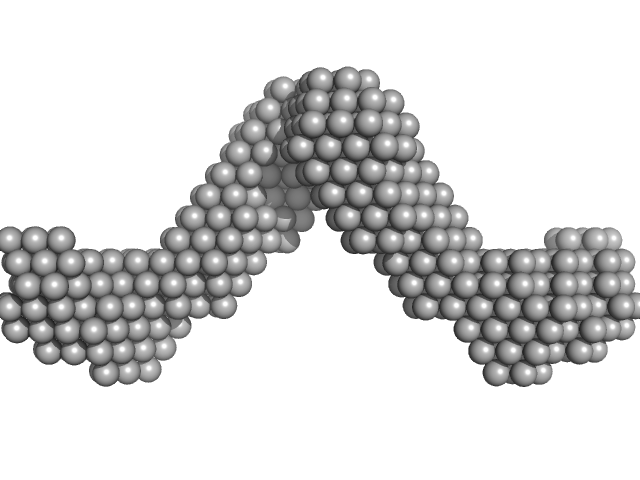
|
| Sample: |
Noelin tetramer, 256 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 6
|
Olfactomedin-1 Has a V-shaped Disulfide-linked Tetrameric Structure.
J Biol Chem 290(24):15092-101 (2015)
Pronker MF, Bos TG, Sharp TH, Thies-Weesie DM, Janssen BJ
|
| RgGuinier |
8.5 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
616 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMX8_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N2) (30% of 9 kDa PEG tails) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMY8_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N3) (60% of 9 kDa PEG tails) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMZ8_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-P3) (60% of 5 kDa PEG tails) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDM29_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N2) encapsulated into brome mosaic virus (BMV) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|