|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMY8_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N3) (60% of 9 kDa PEG tails) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMZ8_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-P3) (60% of 5 kDa PEG tails) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDM29_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N2) encapsulated into brome mosaic virus (BMV) 0, 5000 kDa
|
| Buffer: |
0.05 M Tris-HCl, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDM39_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-P3) encapsulated into brome mosaic virus (BMV) 0, 5000 kDa
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, pH: 4.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDM49_fit1_model1.png)
|
| Sample: |
Iron oxide nanoparticles (NP-N3) encapsulated into hepatitis B virus (HBV) 0, 5000 kDa
|
| Buffer: |
0.5 M LiCl, 50 mM HEPES, 2 mM DTT, pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 23
|
Coat Protein-Dependent Behavior of Poly(ethylene glycol) Tails in Iron Oxide Core Virus-like Nanoparticles.
ACS Appl Mater Interfaces 7(22):12089-98 (2015)
Malyutin AG, Cheng H, Sanchez-Felix OR, Carlson K, Stein BD, Konarev PV, Svergun DI, Dragnea B, Bronstein LM
|
|
|
|
|
|
|

|
| Sample: |
PlaB tetramer, 220 kDa Legionella pneumophila protein
|
| Buffer: |
100 mM Tris 100 mM Nacl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 15
|
Automated pipeline for purification, biophysical and x-ray analysis of biomacromolecular solutions.
Sci Rep 5:10734 (2015)
Graewert MA, Franke D, Jeffries CM, Blanchet CE, Ruskule D, Kuhle K, Flieger A, Schäfer B, Tartsch B, Meijers R, Svergun DI
|
| RgGuinier |
4.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
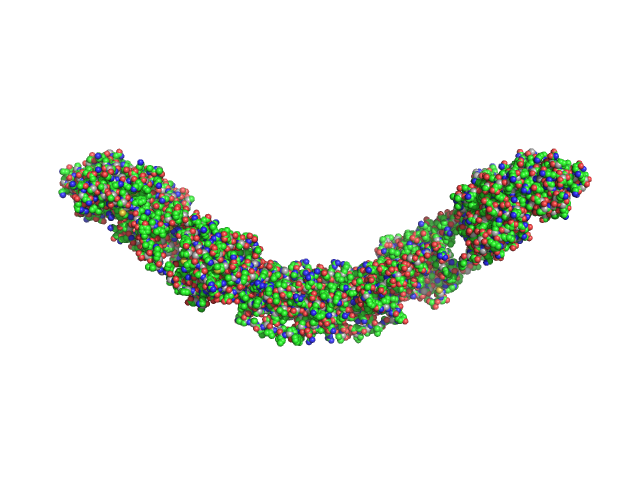
|
| Sample: |
Maltose Binding Protein fused to Protein Interacting with C kinase 1 dimer, 174 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 300 mM NaCl 1 mM maltose 1 mM EGTA 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 13
|
PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner.
Mol Biol Cell 26(7):1308-22 (2015)
Madasu Y, Yang C, Boczkowska M, Bethoney KA, Zwolak A, Rebowski G, Svitkina T, Dominguez R
|
| RgGuinier |
8.4 |
nm |
| Dmax |
27.6 |
nm |
| VolumePorod |
483 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human Filamin A Ig-like domains 20-21* monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 50 mM NaCl 10mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 1
|
Flexible Structure of Peptide-Bound Filamin A Mechanosensor Domain Pair 20-21.
PLoS One 10(8):e0136969 (2015)
Seppälä J, Tossavainen H, Rodic N, Permi P, Pentikäinen U, Ylänne J
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Leishmania braziliensis p23 isoform B monomer, 23 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 5 mM B-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2012 Mar 26
|
Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities.
FEBS J 282(2):388-406 (2015)
Batista FA, Almeida GS, Seraphim TV, Silva KP, Murta SM, Barbosa LR, Borges JC
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Uncharacterized protein monomer, 22 kDa Leishmania braziliensis protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM NaCl, 5 mM B-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2012 Mar 26
|
Identification of two p23 co-chaperone isoforms in Leishmania braziliensis exhibiting similar structures and Hsp90 interaction properties despite divergent stabilities.
FEBS J 282(2):388-406 (2015)
Batista FA, Almeida GS, Seraphim TV, Silva KP, Murta SM, Barbosa LR, Borges JC
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
|
|