|
|
|
|
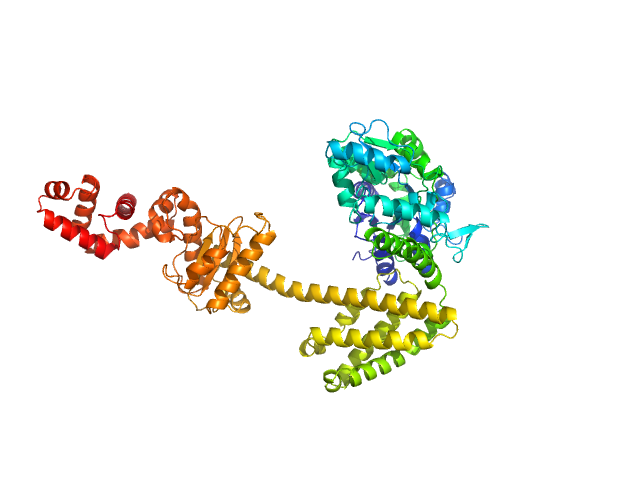
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) monomer, 40 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 monomer, 43 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
|
|
|
|
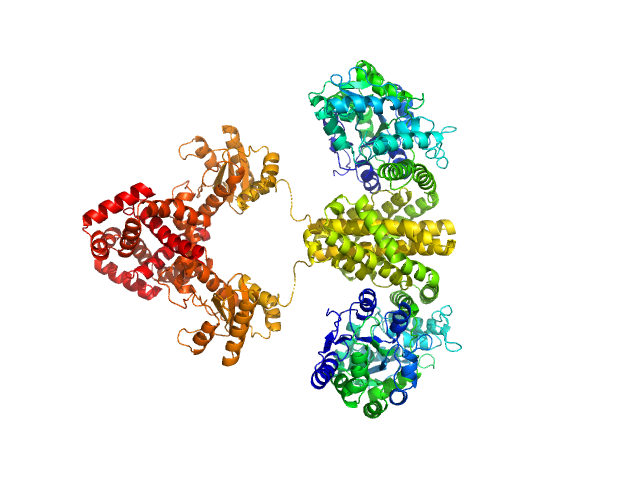
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) dimer, 80 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 dimer, 86 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
345 |
nm3 |
|
|
|
|
|
|
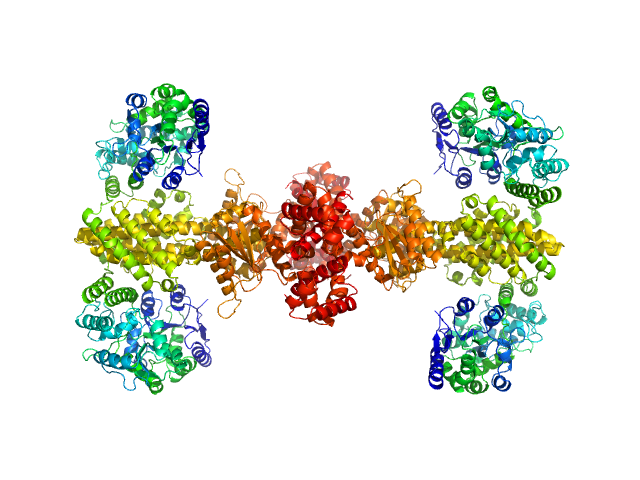
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) tetramer, 161 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 tetramer, 171 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
5.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
826 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDV54_fit1_model1.png)
|
| Sample: |
Phage antirepressor protein Cro dimer, 24 kDa Escherichia coli O157:H7 protein
IR1 22 base pair dsDNA monomer, 14 kDa DNA
|
| Buffer: |
20 mM sodium acetate, 150 mM NaCl, 1 mM TCEP, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 13
|
Structural-function relationship of YdaS, a Cro-type repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7
Marusa Prolic Kalinsek
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
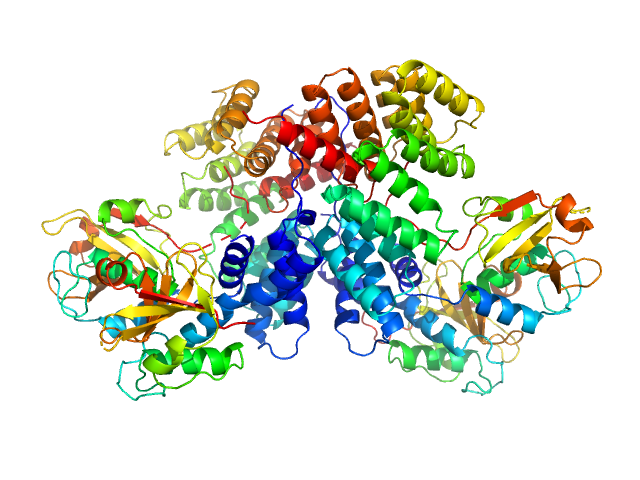
|
| Sample: |
Lipoprotein NlpI dimer, 63 kDa Escherichia coli (strain … protein
Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase tetramer, 73 kDa Escherichia coli (strain … protein
|
| Buffer: |
0.2 M Na/K phosphate pH 7.0, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2023 Sep 7
|
Structural basis for recruitment of peptidoglycan endopeptidase MepS by lipoprotein NlpI.
Nat Commun 15(1):5461 (2024)
Wang S, Huang CH, Lin TS, Yeh YQ, Fan YS, Wang SW, Tseng HC, Huang SJ, Chang YY, Jeng US, Chang CI, Tzeng SR
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alpha domain of Ag43a monomer, 49 kDa Escherichia coli protein
Fragment antigen-binding region Fab10C12 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 3
|
Inhibition of aggregation and biofilm formation by Uropathogenic Escherichia coli
Andrew Whitten
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translocated intimin receptor Tir dimer, 50 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 18
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Translocated intimin receptor Tir dimer, 32 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translocated intimin receptor Tir monomer, 18 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
20 mM Sodium Phosphate, 150 mM NaCl, 1 mM EDTA, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 15
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translocated intimin receptor Tir monomer, 12 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Nov 21
|
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection.
Commun Biol 7(1):179 (2024)
Vieira MFM, Hernandez G, Zhong Q, Arbesú M, Veloso T, Gomes T, Martins ML, Monteiro H, Frazão C, Frankel G, Zanzoni A, Cordeiro TN
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|