|
|
|
|
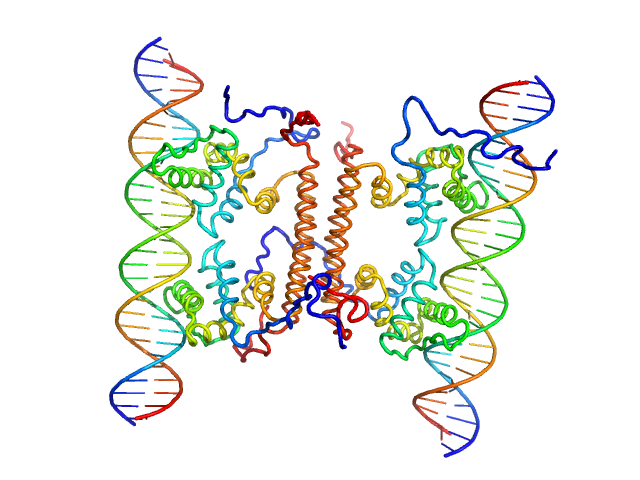
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
Om 30 base pair dsDNA dimer, 37 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 14
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
7.4 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
7.1 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|

|
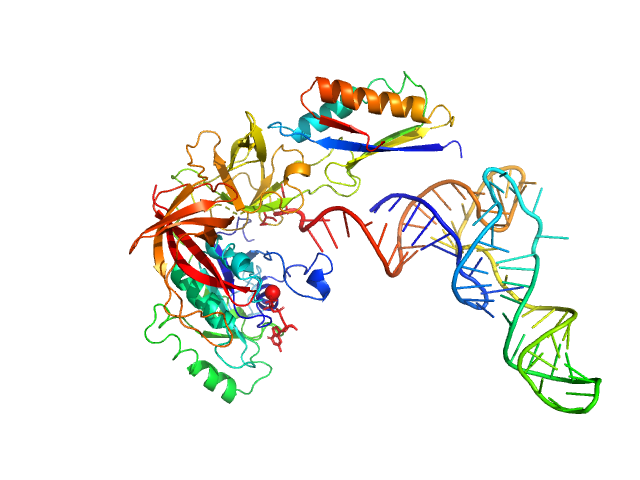
| Sample: |
Transposon Tn3 resolvase dimer, 43 kDa Escherichia coli protein
Tn3 res, resolvase binding site II monomer, 32 kDa DNA
|
| Buffer: |
25 mM Tris, 150 mM NaCl, 10 mM MgCl2, 1% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Nov 6
|
Structural basis for topological regulation of Tn3 resolvase.
Nucleic Acids Res (2022)
Montaño SP, Rowland SJ, Fuller JR, Burke ME, MacDonald AI, Boocock MR, Stark WM, Rice PA
|
| RgGuinier |
5.0 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Translation initiation factor 2 subunit gamma monomer, 46 kDa Saccharolobus solfataricus (strain … protein
Translation initiation factor 2 subunit alpha monomer, 10 kDa Saccharolobus solfataricus (strain … protein
Transfer RNA monomer, 23 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS- NaOH pH 6.7, 200 mM NaCl, 5 mM MgCl 2, 1 mM GDPNP, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (His-tagged) octamer, 565 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
11.0 |
nm |
| Dmax |
32.5 |
nm |
| VolumePorod |
1720 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (His-tagged) octamer, 565 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
8.6 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
830 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 13
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (His-tagged) octamer, 565 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
11.2 |
nm |
| Dmax |
30.5 |
nm |
|
|