|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
58 nucleotide RNA L11-binding domain from E. coli 23S rRNA monomer, 19 kDa Escherichia coli RNA
|
| Buffer: |
10 mM sodium cacodylate, 100 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 9
|
Chaotic advection mixer for capturing transient states of diverse biological macromolecular systems with time-resolved small-angle X-ray scattering
IUCrJ 10(3):363-375 (2023)
Zielinski K, Katz A, Calvey G, Pabit S, Milano S, Aplin C, San Emeterio J, Cerione R, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cell division protein FtsB hexamer, 70 kDa Escherichia coli (strain … protein
Cell division protein FtsL hexamer, 82 kDa Escherichia coli (strain … protein
Cell division protein FtsQ hexamer, 189 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 200 mM NaCl, 0.2% of Cymal-5, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Sep 23
|
Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome
Nature Communications 14(1) (2023)
Nguyen H, Chen X, Parada C, Luo A, Shih O, Jeng U, Huang C, Shih Y, Ma C
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
421 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Colibactin hybrid non-ribosomal peptide synthetase/type I polyketide synthase ClbK dimer, 476 kDa Escherichia coli protein
|
| Buffer: |
50 mM Hepes pH 7.5, 200 mM NaCl, 5 mM DTT, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Sep 14
|
Architecture of a PKS-NRPS hybrid megaenzyme involved in the biosynthesis of the genotoxin colibactin
Structure (2023)
Bonhomme S, Contreras-Martel C, Dessen A, Macheboeuf P
|
| RgGuinier |
8.0 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
1340 |
nm3 |
|
|
|
|
|
|
|
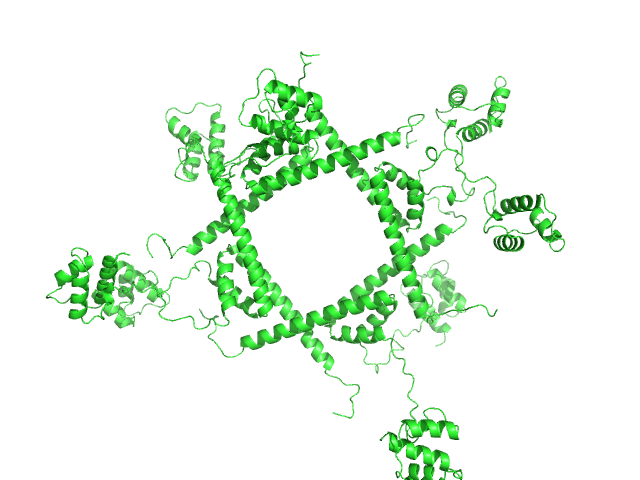
| Sample: |
Phage repressor protein CI (C120S) octamer, 122 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Regulation of the Escherichia coli paaR2-paaA2-ParD2 toxin-antitoxin system
Marusa Prolic Kalinsek
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
428 |
nm3 |
|
|
|
|
|
|
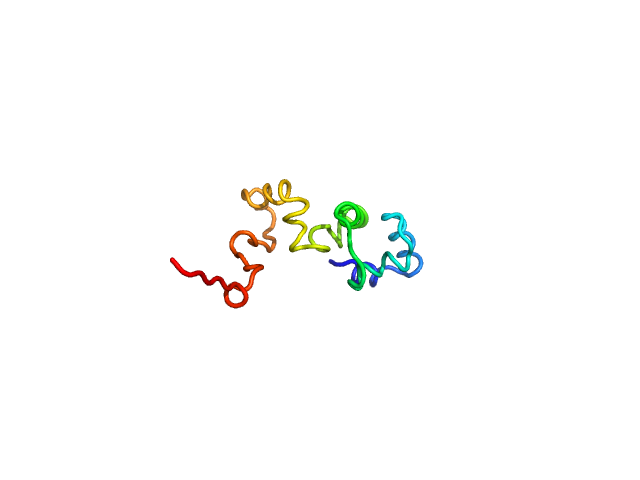
|
| Sample: |
Phage antirepressor protein Cro monomer, 12 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM sodium acetate, 150 mM NaCl, 1 mM TCEP, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 5
|
Structural-function relationship of YdaS, a Cro-type repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7
Marusa Prolic Kalinsek
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Uncharacterized protein YbiB dimer, 75 kDa Escherichia coli (strain … protein
GTPase Obg dimer, 91 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 2 mM DTT, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 24
|
YbiB: a novel interactor of the GTPase ObgE.
Nucleic Acids Res (2023)
Deckers B, Vercauteren S, De Kock V, Martin C, Lazar T, Herpels P, Dewachter L, Verstraeten N, Peeters E, Ballet S, Michiels J, Galicia C, Versées W
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.6 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQB9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQC9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein (mutant: L111N, F118R) monomer, 18 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Apr 14
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQD9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein monomer, 13 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|