|
|
|
|
|
| Sample: |
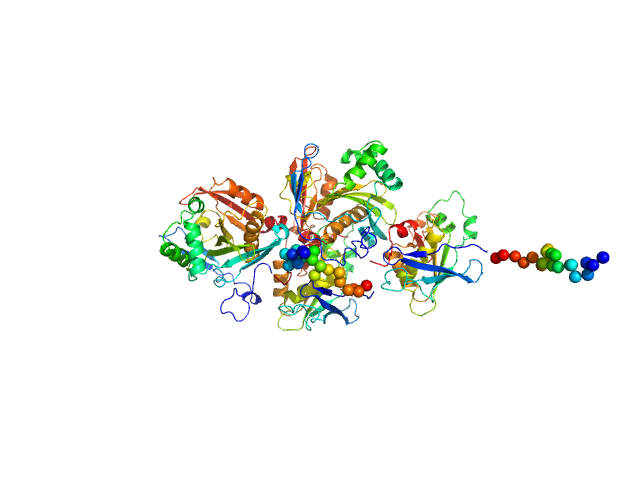
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF tetramer, 223 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Jul 7
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
412 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF monomer, 25 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
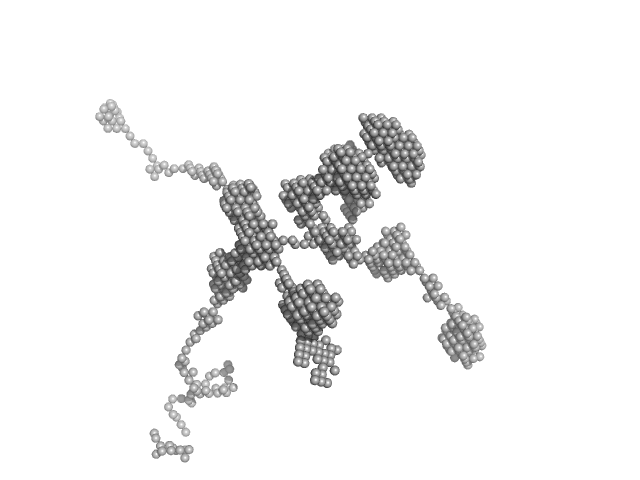
N(5)-hydroxyornithine:cis-anhydromevalonyl coenzyme A-N(5)-transacylase sidF (Δ444-462) dimer, 107 kDa Aspergillus fumigatus (strain … protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2023 Jan 18
|
SidF, a dual substrate N5-acetyl-N5-hydroxy-L-ornithine transacetylase involved in Aspergillus fumigatus siderophore biosynthesis
Journal of Structural Biology: X 11:100119 (2025)
Poonsiri T, Stransky J, Demitri N, Haas H, Cianci M, Benini S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
90.5 |
nm |
| Dmax |
200.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
30.7 |
nm |
| Dmax |
107.4 |
nm |
| VolumePorod |
112000 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Self-amplifying RNA monomer, 3030 kDa RNA
Linear polyethylenimine monomer, 25 kDa none (polymer)
|
| Buffer: |
MBG buffer: 5% w/v D-Glucose, 10mM MES (2-(N-morpholino)ethanesulfonic acid) in double distillated sterile/RNAse free water, pH: 6.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 10
|
Compact polyethylenimine-complexed mRNA molecules as quintessential vaccines
Martin Schroer
|
| RgGuinier |
12.0 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
9100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human immunoglobulin gamma 2 (IgG2) ChiLob 7/4 F(ab')2 - heavy chain T219C/C224S dimer, 52 kDa protein
Human immunoglobulin gamma 2 (IgG2) ChiLob 7/4 F(ab')2 - kappa chain E123C/C214S dimer, 47 kDa protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Apr 12
|
Structure-guided disulfide engineering restricts antibody conformation to elicit TNFR agonism.
Nat Commun 16(1):3495 (2025)
Elliott IG, Fisher H, Chan HTC, Inzhelevskaya T, Mockridge CI, Penfold CA, Duriez PJ, Orr CM, Herniman J, Müller KTJ, Essex JW, Cragg MS, Tews I
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
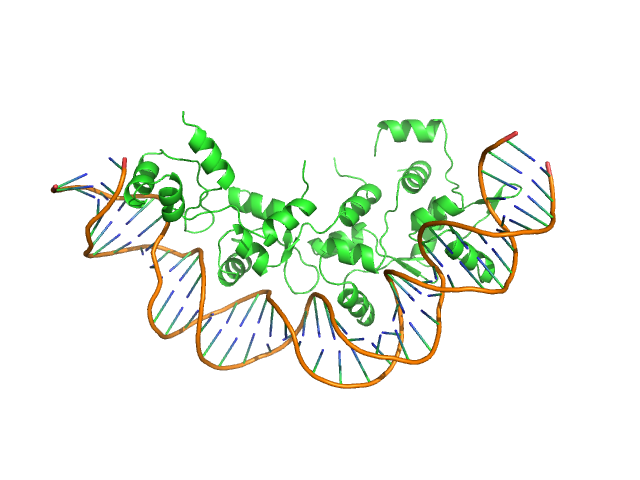
Recombination directionality factor RdfS tetramer, 52 kDa Mesorhizobium japonicum R7A protein
AttP_8 40-mer DNA dimer, 25 kDa DNA
|
| Buffer: |
150 mM Tris-HCl, 300 mM NaCl, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 19
|
Structural basis for control of integrative and conjugative element excision and transfer by the oligomeric winged helix–turn–helix protein RdfS
Nucleic Acids Research 53(6) (2025)
Verdonk C, Agostino M, Eto K, Hall D, Bond C, Ramsay J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
134 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Humanized immunoglobulin G4 monoclonal antibody (lebrikizumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
100 mM glycine-HCl, 200 mM NaCl, pH: 2 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2024 Nov 19
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Humanized immunoglobulin G1 monoclonal antibody (trastuzumab) monomer, 148 kDa Mouse/human protein
|
| Buffer: |
100 mM glycine-HCl, 200 mM NaCl, pH: 2 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 May 20
|
IgG4
and IgG1
undergo common acid‐induced compaction into an alternatively folded state
FEBS Letters (2025)
Imamura H, Honda S
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
240 |
nm3 |
|
|