|
|
|
|
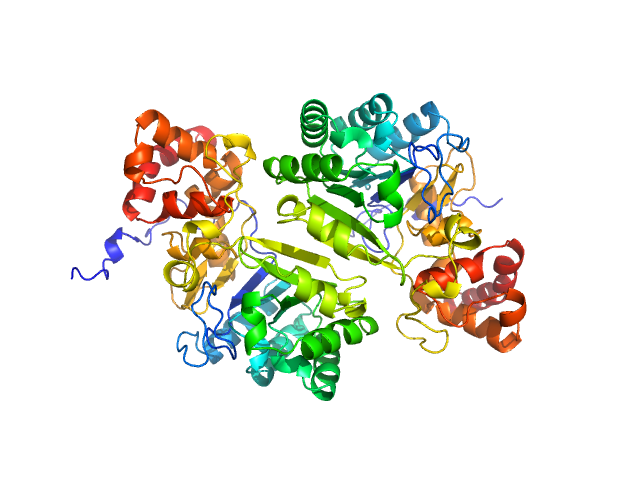
|
| Sample: |
Putative peptide biosynthesis protein YydG dimer, 80 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Nov 29
|
Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat Chem Biol (2023)
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
115 |
nm3 |
|