|
|
|
|

|
| Sample: |
Ferric iron reductase protein FhuF (∆1-17) monomer, 28 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM Phosphate, 200 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 16
|
Conjuring up a ghost: structural and functional characterization of FhuF, a ferric siderophore reductase from E. coli
JBIC Journal of Biological Inorganic Chemistry (2021)
Trindade I, Hernandez G, Lebègue E, Barrière F, Cordeiro T, Piccioli M, Louro R
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
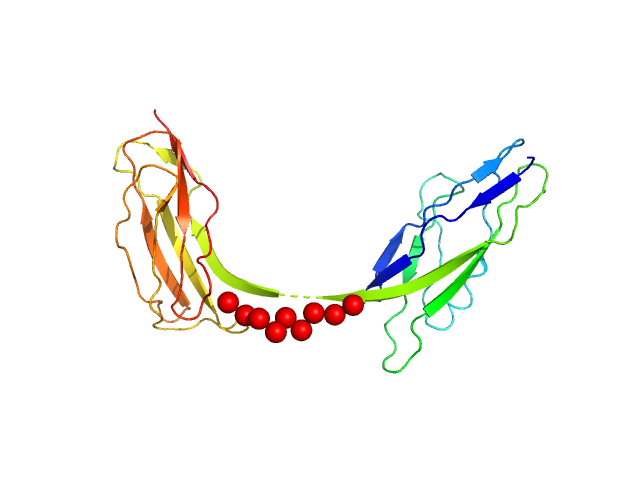
| Sample: |
Intimin, D00-D0 domain (6xHis tagged) monomer, 23 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
10 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 12
|
The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep 10(1):21249 (2020)
Weikum J, Kulakova A, Tesei G, Yoshimoto S, Jægerum LV, Schütz M, Hori K, Skepö M, Harris P, Leo JC, Morth JP
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Intimin (D0-D1 domain, 6xHis tagged) monomer, 23 kDa Escherichia coli O127:H6 … protein
|
| Buffer: |
10 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 12
|
The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep 10(1):21249 (2020)
Weikum J, Kulakova A, Tesei G, Yoshimoto S, Jægerum LV, Schütz M, Hori K, Skepö M, Harris P, Leo JC, Morth JP
|
| RgGuinier |
2.2 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Candidatus Glomeribacter gigasporarum cyclodipeptide synthase monomer, 34 kDa Candidatus Glomeribacter gigasporarum protein
E. coli Phe-tRNAPhe monomer, 25 kDa Escherichia coli RNA
|
| Buffer: |
10 mM MOPS pH6.7; 200 mM NaCl, 8 mM MgCl2, pH: 6.7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Oct 2
|
Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates.
RNA 26(11):1589-1602 (2020)
Bourgeois G, Seguin J, Babin M, Gondry M, Mechulam Y, Schmitt E
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aldehyde-alcohol dehydrogenase dimer, 97 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris 400 mM NaCl 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Nov 27
|
High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):414-421 (2020)
Azmi L, Bragginton EC, Cadby IT, Byron O, Roe AJ, Lovering AL, Gabrielsen M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aldehyde-alcohol dehydrogenase monomer, 48 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
30 mM HEPES, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Jan 30
|
High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):414-421 (2020)
Azmi L, Bragginton EC, Cadby IT, Byron O, Roe AJ, Lovering AL, Gabrielsen M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endonuclease 8 monomer, 31 kDa Escherichia coli protein
|
| Buffer: |
25 mM Bis-Tris, 150 mM NaCl, 2% glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Mar 9
|
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.
Structure (2020)
Eckenroth BE, Cao VB, Averill AM, Dragon JA, Doublié S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 16-mer, 153 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 27
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
8.9 |
nm |
| Dmax |
28.5 |
nm |
| VolumePorod |
410 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 16-mer, 153 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
6.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
336 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha 14-mer, 133 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
5.8 |
nm |
| Dmax |
24.2 |
nm |
| VolumePorod |
308 |
nm3 |
|
|