|
|
|
|

|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
Frataxin, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 Apr 18
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
287 |
nm3 |
|
|
|
|
|
|
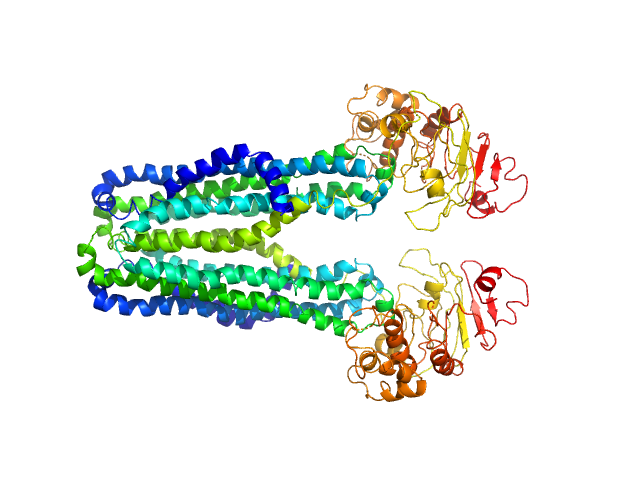
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA dimer, 133 kDa Escherichia coli protein
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at D11, ILL on 2017 Mar 9
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA dimer, 133 kDa Escherichia coli protein
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 1 mM ADP, pH: 7.5 |
| Experiment: |
SANS
data collected at D11, ILL on 2017 Mar 9
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipid A export ATP-binding/permease protein MsbA dimer, 133 kDa Escherichia coli protein
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 8
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
607 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA monomer, 27 kDa Escherichia coli protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 30
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleotide Binding Domain of Lipid A export ATP-binding/permease protein MsbA monomer, 27 kDa Escherichia coli protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 0.5 mM TCEP, 1 mM ADP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 30
|
Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs.
Structure 26(8):1072-1079.e4 (2018)
Josts I, Nitsche J, Maric S, Mertens HD, Moulin M, Haertlein M, Prevost S, Svergun DI, Busch S, Forsyth VT, Tidow H
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Escherichia coli TraE protein (VirB8 homolog) hexamer, 171 kDa Escherichia coli protein
|
| Buffer: |
50 mM sodium phosphate 300 mM NaCl 40 mM imidazole 0.15 % octyl glucose neopentyl glycol (OGNG), pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 2
|
VirB8 homolog TraE from plasmid pKM101 forms a hexameric ring structure and interacts with the VirB6 homolog TraD.
Proc Natl Acad Sci U S A 115(23):5950-5955 (2018)
Casu B, Mary C, Sverzhinsky A, Fouillen A, Nanci A, Baron C
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
360 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Saposin-a monomer, 9 kDa Homo sapiens protein
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine, 47 kDa synthetic construct
Dipeptide and tripeptide permease A monomer, 51 kDa Escherichia coli protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 4
|
Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research.
Structure 26(2):345-355.e5 (2018)
Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
288 |
nm3 |
|
|