|
|
|
|
|
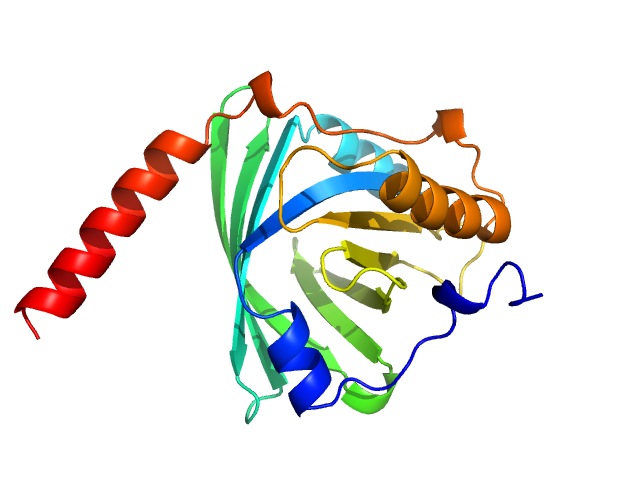
| Sample: |
Alpha-1-acid glycoprotein 1 monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2021 Jan 12
|
SAXS data based glycosylated model of alpha-1-acid glycoprotein
FNU Ashish
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
96 |
nm3 |
|